

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
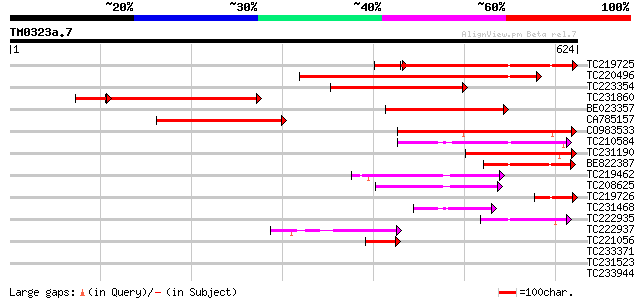
Query= TM0323a.7
(624 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219725 similar to UP|Q9FJY0 (Q9FJY0) Arabidopsis thaliana geno... 332 e-102
TC220496 weakly similar to UP|Q9FJY0 (Q9FJY0) Arabidopsis thalia... 285 3e-77
TC223354 similar to UP|Q9FJY0 (Q9FJY0) Arabidopsis thaliana geno... 254 1e-67
TC231860 similar to UP|Q9FJY0 (Q9FJY0) Arabidopsis thaliana geno... 244 6e-65
BE023357 240 2e-63
CA785157 178 7e-45
CO983533 153 2e-37
TC210584 similar to UP|Q9FMZ5 (Q9FMZ5) Emb|CAB16786.1, partial (... 105 6e-23
TC231190 97 2e-20
BE822387 91 1e-18
TC219462 weakly similar to UP|Q9LHM9 (Q9LHM9) Gb|AAC17092.1, par... 91 2e-18
TC208625 78 1e-14
TC219726 72 7e-13
TC231468 similar to UP|Q9FMZ5 (Q9FMZ5) Emb|CAB16786.1, partial (... 64 2e-10
TC222935 similar to UP|Q8RY64 (Q8RY64) AT4g37080/C7A10_280, part... 60 4e-09
TC222937 similar to UP|Q8RY64 (Q8RY64) AT4g37080/C7A10_280, part... 49 9e-06
TC221056 weakly similar to UP|Q93ZL1 (Q93ZL1) AT5g60720/mup24_13... 45 7e-05
TC233371 weakly similar to UP|Q9M0X9 (Q9M0X9) 4-coumarate-CoA li... 34 0.17
TC231523 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {A... 32 0.84
TC233944 similar to UP|Q9FJV6 (Q9FJV6) Major surface glycoprotei... 32 1.1
>TC219725 similar to UP|Q9FJY0 (Q9FJY0) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K1F13, partial (34%)
Length = 866
Score = 332 bits (851), Expect(2) = e-102
Identities = 162/194 (83%), Positives = 179/194 (91%)
Frame = +3
Query: 431 AFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTK 490
AFLAYGIPQNNVKRVFLLLKAAYNVGG T+SADTIQ+TIL+CRMSRPGQWLR+ FS TK
Sbjct: 183 AFLAYGIPQNNVKRVFLLLKAAYNVGGHTISADTIQNTILKCRMSRPGQWLRLLFSQSTK 362
Query: 491 FRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVR 550
F++GD RQ YA+E EPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRA LG+
Sbjct: 363 FKAGDRRQAYALEQAEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRANLGI- 539
Query: 551 RKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEWIPH 610
RKDQK+L+PKLVE+FTKDS +CP GVM+MI ESLPE LRK+VKKC +LAKSRKCIEWIPH
Sbjct: 540 RKDQKILLPKLVESFTKDSGLCPNGVMDMILESLPEYLRKNVKKC-RLAKSRKCIEWIPH 716
Query: 611 NFTFRYLISKDVLK 624
NFTFRYLISKD++K
Sbjct: 717 NFTFRYLISKDMVK 758
Score = 59.3 bits (142), Expect(2) = e-102
Identities = 27/36 (75%), Positives = 29/36 (80%)
Frame = +2
Query: 402 RLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGI 437
RLEEVD +LKHEEK+AFWINIHNALVMH Y I
Sbjct: 2 RLEEVDXXRLKHEEKIAFWINIHNALVMHVIYYYTI 109
>TC220496 weakly similar to UP|Q9FJY0 (Q9FJY0) Arabidopsis thaliana genomic
DNA, chromosome 5, TAC clone:K1F13, partial (25%)
Length = 836
Score = 285 bits (730), Expect = 3e-77
Identities = 139/266 (52%), Positives = 191/266 (71%)
Frame = +2
Query: 320 SPSSSLSSPSAFSIGDQGDMWSPGFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWI 379
SP S SS + S QG W ++ +SSF+ N FH G +EFSG Y +M+ + +
Sbjct: 41 SPISFPSSGNELSSQSQGSKWGSQWKKHSSFNLNSTNPFHVRGSKEFSGTYCSMIRIQQL 220
Query: 380 YREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQ 439
+ K +IE + ++FRSL+ RLE+V+PR +KHEEKLAFWIN+HN+L MHA L YGI
Sbjct: 221 CTDSQKLKEIEYMLRRFRSLVSRLEDVNPRNMKHEEKLAFWINVHNSLAMHALLIYGISA 400
Query: 440 NNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQT 499
NNVKR+ +LKAAYN+GG T+S D IQ+ IL CR+ RPGQWLR++F S TK + D R+
Sbjct: 401 NNVKRMSSVLKAAYNIGGHTISVDLIQNFILGCRLPRPGQWLRLWFPSMTKPKVRDARKG 580
Query: 500 YAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVP 559
YAI PEPL FALCSG+HSDPAVR+YT KRVF+EL+ AK+EYI++T+ + K+QK+++P
Sbjct: 581 YAIHRPEPLLLFALCSGSHSDPAVRLYTSKRVFEELQCAKEEYIQSTITI-SKEQKIVLP 757
Query: 560 KLVETFTKDSDMCPGGVMEMIQESLP 585
K+V++F K+S + +MEM++ LP
Sbjct: 758 KMVDSFAKNSGLGASDLMEMVKPYLP 835
>TC223354 similar to UP|Q9FJY0 (Q9FJY0) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K1F13, partial (16%)
Length = 455
Score = 254 bits (648), Expect = 1e-67
Identities = 121/150 (80%), Positives = 133/150 (88%)
Frame = +2
Query: 354 LDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICRLEEVDPRKLKH 413
LDN FH EG++EFSGPYSTMVEVSW+YRE K D EKL FRSLICRLEEVDP +LKH
Sbjct: 2 LDNPFHVEGLKEFSGPYSTMVEVSWLYRESQKSADTEKLLLNFRSLICRLEEVDPGRLKH 181
Query: 414 EEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCR 473
EEK+AFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGG T+SADTIQ+TIL+CR
Sbjct: 182 EEKIAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGHTISADTIQNTILKCR 361
Query: 474 MSRPGQWLRMFFSSRTKFRSGDGRQTYAIE 503
MSRPGQWLR+ FS TKF++GD RQ YA+E
Sbjct: 362 MSRPGQWLRLLFSQSTKFKAGDRRQAYALE 451
>TC231860 similar to UP|Q9FJY0 (Q9FJY0) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K1F13, partial (11%)
Length = 787
Score = 244 bits (624), Expect = 6e-65
Identities = 127/173 (73%), Positives = 146/173 (83%), Gaps = 1/173 (0%)
Frame = +2
Query: 106 KPATELIKEIAVLEMEVVYLEQHLLSLYRKAFDQQLSSVSPPTKEESLKYPLTTPRAQVV 165
+PATELI+EIAVLE+EVVYLEQHLLSLYRKAFDQQLSSVSP +KEES+K+PLTT A+ +
Sbjct: 269 QPATELIREIAVLELEVVYLEQHLLSLYRKAFDQQLSSVSPTSKEESVKFPLTTHSARFI 448
Query: 166 KASLPEVLTKEEYPTVQSNDH-ELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGVYRC 224
S+PEVLTK E TVQSN+H +LE +RKE+DR E + RKE + EEKHLDSGV+RC
Sbjct: 449 NFSMPEVLTKRECSTVQSNEHNKLETLRKEYDRYEPETFRKEQSRDLLEEKHLDSGVHRC 628
Query: 225 HSSLSQYSAFTRSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLG 277
HSSLS AFTR SPPA+ LAKSLRACHSQPLSM+EYA+ SSNIISLAEHLG
Sbjct: 629 HSSLSHCPAFTRESPPADSLAKSLRACHSQPLSMLEYAQSSSSNIISLAEHLG 787
Score = 59.7 bits (143), Expect = 4e-09
Identities = 29/40 (72%), Positives = 35/40 (87%)
Frame = +3
Query: 73 RRLQDQFEVRCTLEKALGYKSPSLVNSSEMIMPKPATELI 112
RRLQDQF+VR TLEKALG++S SLVNS+EM++PK AT I
Sbjct: 6 RRLQDQFQVRSTLEKALGFRSSSLVNSNEMMIPKVATSSI 125
>BE023357
Length = 411
Score = 240 bits (612), Expect = 2e-63
Identities = 114/136 (83%), Positives = 123/136 (89%)
Frame = +3
Query: 414 EEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCR 473
EEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYN+GG T+SADTIQ+TIL CR
Sbjct: 3 EEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNIGGHTISADTIQNTILGCR 182
Query: 474 MSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQ 533
+ RPGQW R+FFS RTKF++GDG + Y IE PEPL FALCSGNHSDPAVRVYTPKRV Q
Sbjct: 183 LPRPGQWFRLFFSPRTKFKAGDGHRAYPIERPEPLLLFALCSGNHSDPAVRVYTPKRVLQ 362
Query: 534 ELEVAKDEYIRATLGV 549
ELEVAK+EYIRAT GV
Sbjct: 363 ELEVAKEEYIRATFGV 410
>CA785157
Length = 438
Score = 178 bits (451), Expect = 7e-45
Identities = 98/147 (66%), Positives = 112/147 (75%), Gaps = 4/147 (2%)
Frame = +1
Query: 162 AQVVKASLPEVLTKEEYPTVQSNDHELEAMRKEHDRNELDNLRKECNGGWPEEKHLDSGV 221
A++V+ S PEVLTK VQS DHEL+ ++KE+ E + LRKE N PE KHLDSGV
Sbjct: 1 ARLVEVSKPEVLTKRGSSAVQSIDHELDTLQKEYSGYEHETLRKEYNVHQPEGKHLDSGV 180
Query: 222 YRCHSSLSQYSAFTR--SSPPAEPLAKSLRACHSQPLSMMEYARG--CSSNIISLAEHLG 277
YRCHSSLSQ + FT S+P AE L +SLR CHSQPLSMMEYA+ SS IISLAEHLG
Sbjct: 181 YRCHSSLSQCTTFTTRVSAPAAEVLTESLRTCHSQPLSMMEYAQNVDASSRIISLAEHLG 360
Query: 278 TRISDHVLPVTPNKLSEDMVKCISAIY 304
TRISDH+ P TPN+LSEDMVKCISAIY
Sbjct: 361 TRISDHI-PDTPNRLSEDMVKCISAIY 438
>CO983533
Length = 732
Score = 153 bits (387), Expect = 2e-37
Identities = 80/203 (39%), Positives = 129/203 (63%), Gaps = 7/203 (3%)
Frame = -1
Query: 428 VMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSS 487
VMHA+LAYGIPQ ++KR+ L KAAYN+GG +SA+ I+ I + R G+WL F S+
Sbjct: 732 VMHAYLAYGIPQGSLKRLALFHKAAYNIGGHIISANAIEQAIFCFQTPRIGRWLESFMSA 553
Query: 488 RTKFRSGDGRQ----TYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYI 543
+ ++G+ +Q I + EPL FALC+G SDP ++VYT + ++L +AK ++
Sbjct: 552 ALRKKNGEEKQLIRSKLCITDFEPLVCFALCTGALSDPVLKVYTASNIREQLNIAKRGFL 373
Query: 544 RATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKC---HQLAK 600
+A + V +K KV +PKLVE F++++ + ++ + ES+ + L S++KC K
Sbjct: 372 QANV-VVKKSSKVFLPKLVERFSREASISLHDLLGWVMESVDKKLHDSIQKCLDRKSNKK 196
Query: 601 SRKCIEWIPHNFTFRYLISKDVL 623
S + IEW+P++ FRY+ SKD++
Sbjct: 195 SSQIIEWLPYSSRFRYMFSKDLI 127
>TC210584 similar to UP|Q9FMZ5 (Q9FMZ5) Emb|CAB16786.1, partial (29%)
Length = 648
Score = 105 bits (262), Expect = 6e-23
Identities = 69/194 (35%), Positives = 102/194 (52%), Gaps = 3/194 (1%)
Frame = +2
Query: 428 VMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSS 487
+M+A+L GIP++ V L+ KA NVGG +SA TI+ ILR P W
Sbjct: 8 MMNAYLEKGIPESXEMVVALMHKATINVGGHLLSATTIEHCILRL----PYHW------K 157
Query: 488 RTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATL 547
T + G +TY +E EPL FAL G S PAVR+Y +V ELE+AK EY++A +
Sbjct: 158 FTLSKGGKNHETYGLELSEPLVTFALSCGTWSSPAVRIYRASQVENELEMAKKEYLQAAV 337
Query: 548 GVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRKCIEW 607
G+ K L+PKL++ + D +++ I LP + K K + K+ ++
Sbjct: 338 GI--SISKFLIPKLLDWYLLDFAKDLESLLDWICLQLPSDVGKEAIKFLEKRKTEPISQY 511
Query: 608 I---PHNFTFRYLI 618
+ P+ F FRYL+
Sbjct: 512 VQIMPYEFNFRYLL 553
>TC231190
Length = 553
Score = 97.4 bits (241), Expect = 2e-20
Identities = 48/125 (38%), Positives = 81/125 (64%), Gaps = 3/125 (2%)
Frame = +2
Query: 502 IENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKL 561
+ N +PL FALC+G SDP ++VY+ + +EL +AK E+++A + + +K +KV +PKL
Sbjct: 32 LTNSQPLVCFALCTGALSDPVLKVYSASNISEELNIAKREFLQANV-IVKKSRKVFLPKL 208
Query: 562 VETFTKDSDMCPGGVMEMIQESLPESLRKSVKKCHQLAKSRK---CIEWIPHNFTFRYLI 618
VE F++++ + + + ES+ LR S++KC S+K IEW+P++ FRY+
Sbjct: 209 VERFSREASISVDDLFGWVMESVDRKLRDSMQKCLNPKSSQKPSQIIEWLPYSSRFRYVF 388
Query: 619 SKDVL 623
SKDV+
Sbjct: 389 SKDVI 403
>BE822387
Length = 732
Score = 91.3 bits (225), Expect = 1e-18
Identities = 48/101 (47%), Positives = 71/101 (69%)
Frame = -3
Query: 522 AVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVMEMIQ 581
AVR YT KRV +EL+ AK+EYI++T+ + K K+++PK+V++F K S + +MEM++
Sbjct: 721 AVRXYTSKRVXEELQCAKEEYIQSTITI-SKXXKIVLPKMVDSFAKTSGLGASDLMEMVK 545
Query: 582 ESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLISKDV 622
LP+S RKS+++ Q S K IE PHNFTF YLIS ++
Sbjct: 544 PYLPDSQRKSIQE-FQSKTSWKSIELTPHNFTFHYLISTEL 425
>TC219462 weakly similar to UP|Q9LHM9 (Q9LHM9) Gb|AAC17092.1, partial (5%)
Length = 1212
Score = 90.9 bits (224), Expect = 2e-18
Identities = 57/173 (32%), Positives = 89/173 (50%), Gaps = 5/173 (2%)
Frame = +3
Query: 377 SWIYREGLKWGDIEKLH-----QKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMHA 431
+++Y +G + D +H ++ ++ L+ V+ EEKLAF+IN++N + +HA
Sbjct: 150 AYVYEDGRRI-DYTSIHGSEEFARYLRIVEELQRVEISDSSREEKLAFFINLYNMMAIHA 326
Query: 432 FLAYGIPQNNVKRVFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKF 491
L G P ++R L + Y +GG T S IQ+ ILR P ++ + F
Sbjct: 327 ILVLGHPDGALERRKLFGEFKYVIGGSTYSLSAIQNGILRGNQRPP-------YNLKKPF 485
Query: 492 RSGDGRQTYAIENPEPLSHFALCSGNHSDPAVRVYTPKRVFQELEVAKDEYIR 544
D R T A+ PEPL HFAL G S PA+R Y+P + +EL A ++R
Sbjct: 486 GVKDKRLTVALPYPEPLIHFALVYGTRSGPALRCYSPGNIDEELLDAARNFLR 644
>TC208625
Length = 1218
Score = 77.8 bits (190), Expect = 1e-14
Identities = 47/140 (33%), Positives = 71/140 (50%)
Frame = +1
Query: 403 LEEVDPRKLKHEEKLAFWINIHNALVMHAFLAYGIPQNNVKRVFLLLKAAYNVGGRTVSA 462
L+ V+ +L E LAF+IN++NA+++HA + G + + R Y +GG S
Sbjct: 457 LQRVNLLELSENETLAFFINLYNAMIVHAIIRVGCQEGVINRKSFF-DFHYLIGGHPYSL 633
Query: 463 DTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQTYAIENPEPLSHFALCSGNHSDPA 522
I++ ILR P +S F +GD R +A+ PL HF LC+G S P
Sbjct: 634 GAIKNGILRSNRRSP-------YSLIKPFGTGDRRLEHALVKMNPLVHFGLCNGTKSSPK 792
Query: 523 VRVYTPKRVFQELEVAKDEY 542
VR ++ RV +EL A E+
Sbjct: 793 VRFFSSYRVAEELRSAAREF 852
>TC219726
Length = 363
Score = 72.0 bits (175), Expect = 7e-13
Identities = 35/47 (74%), Positives = 42/47 (88%)
Frame = +1
Query: 578 EMIQESLPESLRKSVKKCHQLAKSRKCIEWIPHNFTFRYLISKDVLK 624
+MI ESLPE LRK+VKKC +LAK R+ +EWIPHNFTFRYLISKD++K
Sbjct: 1 DMILESLPEYLRKNVKKC-RLAKCREGMEWIPHNFTFRYLISKDMVK 138
>TC231468 similar to UP|Q9FMZ5 (Q9FMZ5) Emb|CAB16786.1, partial (21%)
Length = 698
Score = 64.3 bits (155), Expect = 2e-10
Identities = 42/91 (46%), Positives = 50/91 (54%)
Frame = +1
Query: 445 VFLLLKAAYNVGGRTVSADTIQSTILRCRMSRPGQWLRMFFSSRTKFRSGDGRQTYAIEN 504
VFL+ +A NVGG +SA TI+ ILR P W + FS TK R Y +E
Sbjct: 139 VFLVTQATINVGGHVLSATTIEHFILRL----PYHW-KFTFSKGTKNHQMTARSIYGLEL 303
Query: 505 PEPLSHFALCSGNHSDPAVRVYTPKRVFQEL 535
EPL FAL SG S PAVRV T +V +EL
Sbjct: 304 SEPLVTFALSSGTWSSPAVRVDTASQVDKEL 396
>TC222935 similar to UP|Q8RY64 (Q8RY64) AT4g37080/C7A10_280, partial (17%)
Length = 312
Score = 59.7 bits (143), Expect = 4e-09
Identities = 32/103 (31%), Positives = 60/103 (58%), Gaps = 3/103 (2%)
Frame = +1
Query: 519 SDPAVRVYTPKRVFQELEVAKDEYIRATLGVRRKDQKVLVPKLVETFTKDSDMCPGGVME 578
S PAVRVYT +V +ELE AK +Y+ A++G+ K K+++PKL++ + D +++
Sbjct: 1 SSPAVRVYTASKVDEELEAAKRDYLHASVGI-TKTNKLIIPKLLDWYLLDFAKDLESLLD 177
Query: 579 MIQESLPESLRKSVKKCHQLA---KSRKCIEWIPHNFTFRYLI 618
+ LP+ LR +C + + ++ + ++F+FR L+
Sbjct: 178 WVCLQLPDELRNQAVECLERRGRDSLSQMVQMMSYDFSFRLLL 306
>TC222937 similar to UP|Q8RY64 (Q8RY64) AT4g37080/C7A10_280, partial (7%)
Length = 752
Score = 48.5 bits (114), Expect = 9e-06
Identities = 38/149 (25%), Positives = 69/149 (45%), Gaps = 5/149 (3%)
Frame = +1
Query: 288 TPNKLSEDMVKCISAIYCKLA-----DPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMWSP 342
+PN +SE+++KC+S+I +++ D +PP L +P + + + + W P
Sbjct: 355 SPNIISENILKCLSSIILRMSAAKNLDSTADVPP--------LRTPKSKNCVEGIEFWDP 510
Query: 343 GFRNNSSFDARLDNSFHGEGVEEFSGPYSTMVEVSWIYREGLKWGDIEKLHQKFRSLICR 402
S E + GPY + + + + L + + L+ +
Sbjct: 511 -------------YSICLEFGKRDIGPYKQLRSIETKSFDPKRTAKSLFLLHRLKLLLRK 651
Query: 403 LEEVDPRKLKHEEKLAFWINIHNALVMHA 431
L V+ L H+EKLAFWINI+N+ +M+A
Sbjct: 652 LACVNIENLNHQEKLAFWINIYNSCMMNA 738
>TC221056 weakly similar to UP|Q93ZL1 (Q93ZL1) AT5g60720/mup24_130, partial
(15%)
Length = 1022
Score = 45.4 bits (106), Expect = 7e-05
Identities = 19/39 (48%), Positives = 29/39 (73%)
Frame = +1
Query: 392 LHQKFRSLICRLEEVDPRKLKHEEKLAFWINIHNALVMH 430
L +K R L+ L+ VD + L +++KLAFWIN++NA +MH
Sbjct: 901 LLRKLRILMSNLQTVDLKSLTNQQKLAFWINVYNACIMH 1017
>TC233371 weakly similar to UP|Q9M0X9 (Q9M0X9) 4-coumarate-CoA ligase-like
protein, partial (45%)
Length = 943
Score = 34.3 bits (77), Expect = 0.17
Identities = 28/98 (28%), Positives = 41/98 (41%)
Frame = +3
Query: 231 YSAFTRSSPPAEPLAKSLRACHSQPLSMMEYARGCSSNIISLAEHLGTRISDHVLPVTPN 290
+S T +SP + +K+ H + S+I +L +HL R S+H P P
Sbjct: 111 FSQMTPTSPSSHSSSKAFPLSHPKS----------PSSIPTLPKHLPWRTSNHRSPNLPT 260
Query: 291 KLSEDMVKCISAIYCKLADPPMTLPPGLLSPSSSLSSP 328
S ++ YC L TL L P S+ SSP
Sbjct: 261 VSSN*A*TKTTSSYCSLPTASTTLYASSLPPPSAPSSP 374
>TC231523 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Arabidopsis
thaliana;} , partial (32%)
Length = 878
Score = 32.0 bits (71), Expect = 0.84
Identities = 27/82 (32%), Positives = 38/82 (45%), Gaps = 12/82 (14%)
Frame = +1
Query: 135 KAFDQQLSSVSPPTK----EESLKYPLTTPRAQVVKASLPEVLT-KEEYPTVQ------- 182
+ D QLS V TK + S+K+P + V S P+ LT K + P
Sbjct: 172 RTHDNQLSYVGGDTKILAVDRSVKFPAFLSKLAAVCDSAPQDLTFKYQLPGEDLDALISV 351
Query: 183 SNDHELEAMRKEHDRNELDNLR 204
+ND +LE M E+DR NL+
Sbjct: 352 TNDDDLEHMMHEYDRLYRPNLK 417
>TC233944 similar to UP|Q9FJV6 (Q9FJV6) Major surface glycoprotein-like,
partial (24%)
Length = 428
Score = 31.6 bits (70), Expect = 1.1
Identities = 17/35 (48%), Positives = 20/35 (56%)
Frame = +1
Query: 306 KLADPPMTLPPGLLSPSSSLSSPSAFSIGDQGDMW 340
KL +PPMT PPGL S + L+ P F GD W
Sbjct: 118 KLGEPPMTFPPGLPS-CNPLADPPVF-----GDCW 204
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,346,753
Number of Sequences: 63676
Number of extensions: 411448
Number of successful extensions: 2462
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 2399
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2434
length of query: 624
length of database: 12,639,632
effective HSP length: 103
effective length of query: 521
effective length of database: 6,081,004
effective search space: 3168203084
effective search space used: 3168203084
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0323a.7


