

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
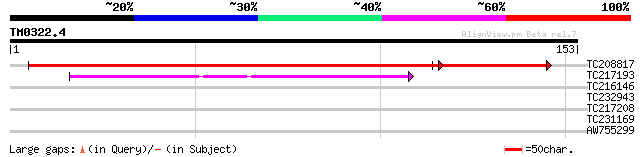
Query= TM0322.4
(153 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC208817 similar to GB|AAP68295.1|31711878|BT008856 At5g37370 {A... 205 1e-60
TC217193 similar to UP|Q7XJN2 (Q7XJN2) At2g40650 protein, partia... 42 9e-05
TC216146 similar to UP|Q7Y256 (Q7Y256) Beta-cyanoalanine synthas... 32 0.12
TC232943 similar to UP|OLE2_ORYSA (Q40646) Oleosin 18 kDa (OSE72... 28 1.7
TC217208 similar to UP|Q9M090 (Q9M090) Kinase binding protein-li... 27 5.0
TC231169 26 8.5
AW755299 26 8.5
>TC208817 similar to GB|AAP68295.1|31711878|BT008856 At5g37370 {Arabidopsis
thaliana;} , partial (53%)
Length = 838
Score = 205 bits (522), Expect(2) = 1e-60
Identities = 99/112 (88%), Positives = 101/112 (89%)
Frame = +1
Query: 6 IMEIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNC 65
IMEIQT GRPI+SLLEKVLCMNILSSDYFKELYRLKTY EVIDEIYNQVDHVEPWM GNC
Sbjct: 190 IMEIQTCGRPIDSLLEKVLCMNILSSDYFKELYRLKTYHEVIDEIYNQVDHVEPWMTGNC 369
Query: 66 RGPSTFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDPNTV 117
RGPST FCLLY FFTM LTVKQMHGLLKH DSPYIR VGFLYLRY DP T+
Sbjct: 370 RGPSTAFCLLYKFFTMKLTVKQMHGLLKHLDSPYIRAVGFLYLRYCADPKTL 525
Score = 43.9 bits (102), Expect(2) = 1e-60
Identities = 23/32 (71%), Positives = 26/32 (80%)
Frame = +3
Query: 115 NTV*LV*TIYKG**GIFSWI*WTNDYNGCIYP 146
+T+ LV* I K **GIFS I*W+NDYNGCI P
Sbjct: 519 DTMELV*AICKR**GIFSRI*WSNDYNGCICP 614
>TC217193 similar to UP|Q7XJN2 (Q7XJN2) At2g40650 protein, partial (88%)
Length = 1691
Score = 42.4 bits (98), Expect = 9e-05
Identities = 27/93 (29%), Positives = 49/93 (52%)
Frame = +3
Query: 17 ESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPWMAGNCRGPSTFFCLLY 76
++L+EK+L I + Y+KE T ++D+ ++DH+ GN R P+ F CL+
Sbjct: 261 QNLVEKILRSKIYQNTYWKEQCFGLTAETLVDKAM-ELDHLGGTYGGN-RKPTPFMCLVM 434
Query: 77 *FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLR 109
+ + + +K+ D Y+R++G YLR
Sbjct: 435 KMLQIQPEKEIVIEFIKNEDYKYVRILGAFYLR 533
>TC216146 similar to UP|Q7Y256 (Q7Y256) Beta-cyanoalanine synthase, partial
(96%)
Length = 1431
Score = 32.0 bits (71), Expect = 0.12
Identities = 15/27 (55%), Positives = 17/27 (62%)
Frame = -2
Query: 91 LLKHPDSPYIRVVGFLYLRYVPDPNTV 117
LL +PYI +GFL LRY P P TV
Sbjct: 938 LLSLGSTPYILTLGFLDLRYCPTPETV 858
>TC232943 similar to UP|OLE2_ORYSA (Q40646) Oleosin 18 kDa (OSE721), partial
(8%)
Length = 444
Score = 28.1 bits (61), Expect = 1.7
Identities = 10/22 (45%), Positives = 17/22 (76%)
Frame = +2
Query: 16 IESLLEKVLCMNILSSDYFKEL 37
+E+++++ LC NIL +Y KEL
Sbjct: 248 VEAIIQQCLCENILQLEYIKEL 313
>TC217208 similar to UP|Q9M090 (Q9M090) Kinase binding protein-like, partial
(60%)
Length = 1498
Score = 26.6 bits (57), Expect = 5.0
Identities = 10/22 (45%), Positives = 14/22 (63%)
Frame = +1
Query: 93 KHPDSPYIRVVGFLYLRYVPDP 114
+HP PY+ +G+LY R P P
Sbjct: 181 RHPLRPYLDYIGYLYQRMDPLP 246
>TC231169
Length = 884
Score = 25.8 bits (55), Expect = 8.5
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 4/34 (11%)
Frame = -2
Query: 35 KELY-RLKTYLEVIDE---IYNQVDHVEPWMAGN 64
+ELY RL Y +++ + Y+Q +H PW G+
Sbjct: 385 RELYPRLSVYNQIVGQKVYSYHQYNHPHPWQEGS 284
>AW755299
Length = 427
Score = 25.8 bits (55), Expect = 8.5
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 4/34 (11%)
Frame = -2
Query: 35 KELY-RLKTYLEVIDE---IYNQVDHVEPWMAGN 64
+ELY RL Y +++ + Y+Q +H PW G+
Sbjct: 423 RELYPRLSVYNQIVGQKVYSYHQYNHPHPWQEGS 322
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.352 0.162 0.599
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,464,773
Number of Sequences: 63676
Number of extensions: 104530
Number of successful extensions: 773
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 768
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 772
length of query: 153
length of database: 12,639,632
effective HSP length: 89
effective length of query: 64
effective length of database: 6,972,468
effective search space: 446237952
effective search space used: 446237952
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0322.4


