

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0322.12
(601 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
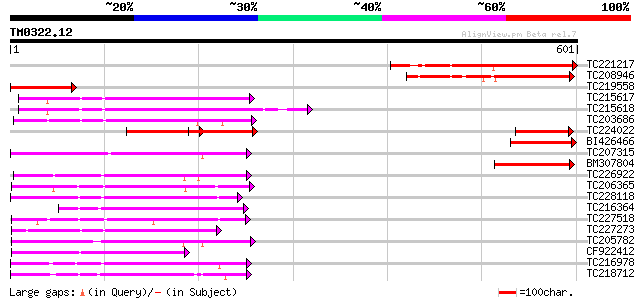
Score E
Sequences producing significant alignments: (bits) Value
TC221217 similar to UP|Q8SA64 (Q8SA64) NIMA-related protein kina... 222 3e-58
TC208946 similar to UP|Q8SA64 (Q8SA64) NIMA-related protein kina... 150 2e-36
TC219558 similar to UP|Q8SA64 (Q8SA64) NIMA-related protein kina... 131 1e-30
TC215617 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase ... 130 2e-30
TC215618 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase ... 126 2e-29
TC203686 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, complete 126 3e-29
TC224022 homologue to UP|Q9LXP3 (Q9LXP3) Protein kinase-like pro... 125 7e-29
BI426466 similar to GP|19699307|gb| AT5g28290/T8M17_60 {Arabidop... 121 8e-28
TC207315 UP|Q8H1P2 (Q8H1P2) Phosphoenolpyruvate carboxylase kina... 120 1e-27
BM307804 similar to GP|18766642|gb| NIMA-related protein kinase ... 115 7e-26
TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%) 113 2e-25
TC206365 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, parti... 111 1e-24
TC228118 similar to UP|KP19_ARATH (Q39030) Serine/threonine-prot... 108 7e-24
TC216364 similar to UP|Q9MAI7 (Q9MAI7) F12M16.4, partial (58%) 107 2e-23
TC227518 similar to UP|Q93VD3 (Q93VD3) At1g30270/F12P21_6 (CBL-i... 106 3e-23
TC227273 weakly similar to UP|KGS9_YEAST (P43637) Probable serin... 106 3e-23
TC205782 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, pa... 106 3e-23
CF922412 105 4e-23
TC216978 homologue to UP|Q8RXH5 (Q8RXH5) Osmotic stress-activate... 104 1e-22
TC218712 similar to UP|Q40264 (Q40264) Protein kinase , partial ... 104 1e-22
>TC221217 similar to UP|Q8SA64 (Q8SA64) NIMA-related protein kinase, partial
(20%)
Length = 670
Score = 222 bits (566), Expect = 3e-58
Identities = 134/207 (64%), Positives = 147/207 (70%), Gaps = 9/207 (4%)
Frame = +3
Query: 404 SHAPSVKFPPPTRRASHPLPTRASMAKATLYTNVGTFPRVDS-DVSVNAPQIDKIAEFPL 462
SHAPS KF P R + P LYTNV +DS DVS+NAP+IDKI EFP+
Sbjct: 9 SHAPSRKFSTPPRTRARATPN--------LYTNV--LGSLDSLDVSINAPRIDKIVEFPM 158
Query: 463 ASCEDLPFFPVHVPSSTSVHCYSSSAGS-AYRSITKEKC-IHEDKVTVPGG-----SCPK 515
A CED PF P+ PSSTS C SSSAGS A SITK+KC I EDKVT+P PK
Sbjct: 159 AFCED-PFSPIRGPSSTSARCSSSSAGSTADCSITKDKCTIQEDKVTLPTSITDACPAPK 335
Query: 516 GSECS-KHVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVL 574
++CS +HV VS HSS +L QRRFDTSSYQQRAEALEGLLEFSARLLQ QRFDELGVL
Sbjct: 336 ETKCSYEHVKDCVSSHSSTDLDQRRFDTSSYQQRAEALEGLLEFSARLLQQQRFDELGVL 515
Query: 575 LKPFGLEKVSPRETAIWLAKSFKETVI 601
LKPFGLEKVSPRETAIWL KSF T +
Sbjct: 516 LKPFGLEKVSPRETAIWLTKSFXXTAV 596
>TC208946 similar to UP|Q8SA64 (Q8SA64) NIMA-related protein kinase, partial
(18%)
Length = 1012
Score = 150 bits (378), Expect = 2e-36
Identities = 97/201 (48%), Positives = 123/201 (60%), Gaps = 23/201 (11%)
Frame = +3
Query: 421 PLPTRASMAKATLYTNVGTFPRVDS-DVSVNAPQIDKIAEFPLASCEDLPFFPVHVPSST 479
PLPTR +++ +V V S D+SVN+P+IDK+AEFPL S ED FP+ SS+
Sbjct: 21 PLPTRGGISQPPC-RSVSLLSHVSSPDISVNSPRIDKMAEFPLTSYED-SLFPIKRASSS 194
Query: 480 SVHCYSSSAGSAY--RSITKEKC---IHEDKVTVPGGSC-----------------PKGS 517
+ SS +Y S +KC +++ PG C K
Sbjct: 195 A---RGSSGFPSYSNHSTVIDKCTVEVYDTASNKPG--CMDAWQGIKQSMLKEIDEDKSG 359
Query: 518 ECSKHVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKP 577
++ TAG S H+S++L +R+FD SS+QQRAEALEGLLEFSARLLQ R+DELGVLLKP
Sbjct: 360 SSDQNATAGASSHTSSDLRRRQFDPSSFQQRAEALEGLLEFSARLLQQARYDELGVLLKP 539
Query: 578 FGLEKVSPRETAIWLAKSFKE 598
FG KVSPRETAIWL+KSFKE
Sbjct: 540 FGPGKVSPRETAIWLSKSFKE 602
>TC219558 similar to UP|Q8SA64 (Q8SA64) NIMA-related protein kinase, partial
(11%)
Length = 455
Score = 131 bits (329), Expect = 1e-30
Identities = 63/71 (88%), Positives = 69/71 (96%)
Frame = +1
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYE+LEQIGKG+FGSALLVRHKHEKK YVLKKIRLARQ++R+RRSAH EMELISK+RN
Sbjct: 241 MEQYEILEQIGKGAFGSALLVRHKHEKKKYVLKKIRLARQTERSRRSAHLEMELISKLRN 420
Query: 61 PFIVEYKDSWV 71
PFIVEYKDSWV
Sbjct: 421 PFIVEYKDSWV 453
>TC215617 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase kinase
kinase alpha, partial (51%)
Length = 1451
Score = 130 bits (327), Expect = 2e-30
Identities = 79/255 (30%), Positives = 138/255 (53%), Gaps = 5/255 (1%)
Frame = +3
Query: 10 IGKGSFGSALLVRHKHEKKLYVLKKIRLA---RQSDRTRRSAHQEMELISKVRNPFIVEY 66
+G+G+FG L + +L +K++R+ + S + +QE+ L+S++ +P IV+Y
Sbjct: 672 LGRGTFGHVYLGFNSDSGQLCAIKEVRVVCDDQSSKECLKQLNQEIHLLSQLSHPNIVQY 851
Query: 67 KDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVNHIL 126
S + + + + + Y G + +++ F E + + Q++ L YLH + +
Sbjct: 852 YGSDLGEET-LSVYLEYVSGGSIHKLLQEYGA--FKEPVIQNYTRQIVSGLSYLHGRNTV 1022
Query: 127 HRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADI-PYGSKSD 185
HRD+K +NI + N +I+L DFG+AK + S S G+P +M PE++ + Y D
Sbjct: 1023HRDIKGANILVDPNGEIKLADFGMAKHINSSSSMLSFKGSPYWMAPEVVMNTNGYSLPVD 1202
Query: 186 IWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAP-LPTMYSSAFRGLVKSMLRKNPE 244
IWSLGC I EMA KP + ++ A I KI S P +P SS + ++ L+++P
Sbjct: 1203IWSLGCTILEMATSKPPWNQYEGAAAIFKIGNSRDMPEIPDHLSSDAKNFIQLCLQRDPS 1382
Query: 245 LRPPASELLNHPHLQ 259
RP A +L+ HP ++
Sbjct: 1383ARPTAQKLIEHPFIR 1427
>TC215618 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase kinase
kinase alpha, partial (73%)
Length = 2291
Score = 126 bits (317), Expect = 2e-29
Identities = 90/318 (28%), Positives = 160/318 (50%), Gaps = 6/318 (1%)
Frame = +1
Query: 10 IGKGSFGSALLVRHKHEKKLYVLKKIRLA---RQSDRTRRSAHQEMELISKVRNPFIVEY 66
+G+G+FG L + ++ +K++++ S + +QE+ L++++ +P IV+Y
Sbjct: 634 LGRGTFGHVYLGFNSENGQMCAIKEVKVVFDDHTSKECLKQLNQEINLLNQLSHPNIVQY 813
Query: 67 KDS-WVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVNHI 125
S VE+ + + + Y G + +++ F E + + Q++ L YLH +
Sbjct: 814 HGSELVEES--LSVYLEYVSGGSIHKLLQEYGP--FKEPVIQNYTRQIVSGLAYLHGRNT 981
Query: 126 LHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADI-PYGSKS 184
+HRD+K +NI + N +I+L DFG+AK + S S G+P +M PE++ + Y
Sbjct: 982 VHRDIKGANILVDPNGEIKLADFGMAKHINSSASMLSFKGSPYWMAPEVVMNTNGYSLPV 1161
Query: 185 DIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAP-LPTMYSSAFRGLVKSMLRKNP 243
DIWSLGC I EMA KP + ++ A I KI S P +P S+ + +K L+++P
Sbjct: 1162DIWSLGCTIIEMATSKPPWNQYEGVAAIFKIGNSKDMPEIPEHLSNDAKKFIKLCLQRDP 1341
Query: 244 ELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFVVPDSVSTLSD 303
RP A +LL+HP ++ + S R FP + S P + + S+
Sbjct: 1342LARPTAQKLLDHPFIRDQSATKAANV-SITRDAFPCMFDGSR--------TPPVLESHSN 1494
Query: 304 QDKCLSLSNDRSLNPSIS 321
+ SL D + P+++
Sbjct: 1495RTSITSLDGDYASKPALA 1548
>TC203686 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, complete
Length = 1502
Score = 126 bits (316), Expect = 3e-29
Identities = 84/269 (31%), Positives = 146/269 (54%), Gaps = 12/269 (4%)
Frame = +2
Query: 5 EVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFIV 64
+V++ +GKG+ G LV+HK + + LK I++ + + R+ QE+++ + + P++V
Sbjct: 299 DVIKVVGKGNGGVVQLVQHKWTSQFFALKVIQMNIE-ESMRKQIAQELKINQQAQCPYVV 475
Query: 65 EYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLH-VN 123
S+ E G + II+ Y + G LAD +KK + PE+ L Q+L L YLH
Sbjct: 476 VCYQSFYENGV-ISIILEYMDGGSLADLLKKVKTI--PEDYLAAICKQVLKGLVYLHHEK 646
Query: 124 HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTS-DDLASSIVGTPSYMCPELL--ADIPY 180
HI+HRD+K SN+ + ++++ DFG++ ++ S A++ +GT +YM PE + + Y
Sbjct: 647 HIIHRDLKPSNLLINHIGEVKITDFGVSAIMESTSGQANTFIGTYNYMSPERINGSQRGY 826
Query: 181 GSKSDIWSLGCCIYEMA----ALKPAFKAFDIQALINKINKSIVAPLP----TMYSSAFR 232
KSDIWSLG + E A P ++ +++ I + P P +S+ F
Sbjct: 827 NYKSDIWSLGLILLECALGRFPYAPPDQSETWESIFELIETIVDKPPPIPPSEQFSTEFC 1006
Query: 233 GLVKSMLRKNPELRPPASELLNHPHLQPY 261
+ + L+K+P+ R A EL+ HP + Y
Sbjct: 1007SFISACLQKDPKDRLSAQELMAHPFVNMY 1093
>TC224022 homologue to UP|Q9LXP3 (Q9LXP3) Protein kinase-like protein,
partial (19%)
Length = 937
Score = 125 bits (313), Expect = 7e-29
Identities = 58/82 (70%), Positives = 67/82 (80%)
Frame = +2
Query: 125 ILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPYGSKS 184
+LHRD+KCSNIFLTK++D+RLGDFGLAK L +DDLASS+VGTP+YMCPELLADIPYG KS
Sbjct: 5 VLHRDLKCSNIFLTKDRDVRLGDFGLAKTLKADDLASSVVGTPNYMCPELLADIPYGFKS 184
Query: 185 DIWSLGCCIYEMAALKPAFKAF 206
DIWSLG P F +F
Sbjct: 185 DIWSLGLLYIRDGCSSPCF*SF 250
Score = 106 bits (265), Expect = 3e-23
Identities = 48/73 (65%), Positives = 59/73 (80%)
Frame = +3
Query: 190 GCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRKNPELRPPA 249
GCCIYEMAA +PAFKAFD+ LI+KIN+S + PLP YS + + L+K MLRKNPE RP A
Sbjct: 201 GCCIYEMAAHRPAFKAFDMAGLISKINRSSIGPLPPCYSPSLKTLIKGMLRKNPEHRPTA 380
Query: 250 SELLNHPHLQPYI 262
SE+L HP+LQPY+
Sbjct: 381 SEVLKHPYLQPYL 419
Score = 84.3 bits (207), Expect = 1e-16
Identities = 43/61 (70%), Positives = 48/61 (78%)
Frame = +2
Query: 537 QRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKSF 596
QR D S++QRAEALE LLE SA LLQ R +EL V+LKPFG +KVSPRETAIWLAKS
Sbjct: 542 QR*LDVKSFRQRAEALEELLELSAELLQQNRLEELQVVLKPFGKDKVSPRETAIWLAKSL 721
Query: 597 K 597
K
Sbjct: 722 K 724
>BI426466 similar to GP|19699307|gb| AT5g28290/T8M17_60 {Arabidopsis
thaliana}, partial (11%)
Length = 420
Score = 121 bits (304), Expect = 8e-28
Identities = 62/70 (88%), Positives = 65/70 (92%)
Frame = +2
Query: 531 SSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAI 590
SSAE + RFDTSSYQQRA+ALEGLLEFSARLLQ QRF+ELGVLLKPFG EKVSPRETAI
Sbjct: 2 SSAESRKHRFDTSSYQQRAKALEGLLEFSARLLQQQRFEELGVLLKPFGPEKVSPRETAI 181
Query: 591 WLAKSFKETV 600
WLAKSFKETV
Sbjct: 182 WLAKSFKETV 211
>TC207315 UP|Q8H1P2 (Q8H1P2) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1063
Score = 120 bits (302), Expect = 1e-27
Identities = 78/259 (30%), Positives = 126/259 (48%), Gaps = 4/259 (1%)
Frame = +3
Query: 2 EQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVR-N 60
E+Y+VLE++G+G FG+ H+ K Y K I R + RR E + +S + +
Sbjct: 51 EEYQVLEELGRGRFGTVFRCFHRTSNKFYAAKLIEKRRLLNEDRRCIEMEAKAMSFLSPH 230
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P I++ D++ E I++ C+ L D I + P L QLL A+ +
Sbjct: 231 PNILQIMDAF-EDADSCSIVLELCQPHTLLDRIAAQGPLTEPHA--ASLLKQLLEAVAHC 401
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPY 180
H + HRD+K NI + ++L DFG A+ L S +VGTP Y+ PE++ Y
Sbjct: 402 HAQGLAHRDIKPENILFDEGNKLKLSDFGSAEWLGEGSSMSGVVGTPYYVAPEVIMGREY 581
Query: 181 GSKSDIWSLGCCIYEMAALKPAF---KAFDIQALINKINKSIVAPLPTMYSSAFRGLVKS 237
K D+WS G +Y M A P F A +I + + N + + + S+ + L++
Sbjct: 582 DEKVDVWSSGVILYAMLAGFPPFYGESAPEIFESVLRANLRFPSLIFSSVSAPAKDLLRK 761
Query: 238 MLRKNPELRPPASELLNHP 256
M+ ++P R A + L HP
Sbjct: 762 MISRDPSNRISAHQALRHP 818
>BM307804 similar to GP|18766642|gb| NIMA-related protein kinase {Populus x
canescens}, partial (13%)
Length = 422
Score = 115 bits (287), Expect = 7e-26
Identities = 56/84 (66%), Positives = 68/84 (80%)
Frame = +3
Query: 515 KGSECSKHVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVL 574
K ++ T G S H+S++L +R+FDTSS++QRAEALEGLLEFSARLLQ +R+ ELGVL
Sbjct: 117 KSGSSDQNATTGASSHNSSDLRRRQFDTSSFRQRAEALEGLLEFSARLLQQERYGELGVL 296
Query: 575 LKPFGLEKVSPRETAIWLAKSFKE 598
LKPFG K SPRETAIWL+KS KE
Sbjct: 297 LKPFGPGKASPRETAIWLSKSLKE 368
>TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%)
Length = 1190
Score = 113 bits (283), Expect = 2e-25
Identities = 85/263 (32%), Positives = 127/263 (47%), Gaps = 11/263 (4%)
Frame = +1
Query: 5 EVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN-PFI 63
E L +G G+ G+ VRHK Y LK I + R RR A E ++ +V + P +
Sbjct: 220 EKLAILGHGNGGTVYKVRHKATSATYALKIIHSDTDATRRRR-ALSETSILRRVTDCPHV 396
Query: 64 VEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVN 123
V + S+ + V I++ Y + G L A+ + F EE+L K +L L YLH
Sbjct: 397 VRFHSSFEKPSGDVAILMEYMDGGTLETALAASG--TFSEERLAKVARDVLEGLAYLHAR 570
Query: 124 HILHRDVKCSNIFLTKNQDIRLGDFGLAK-MLTSDDLASSIVGTPSYMCPELLADIPYGS 182
+I HRD+K +NI + D+++ DFG++K M S + +S VGT +YM P+ YG
Sbjct: 571 NIAHRDIKPANILVNSEGDVKIADFGVSKLMCRSLEACNSYVGTCAYMSPDRFDPEAYGG 750
Query: 183 K-----SDIWSLGCCIYEMAA----LKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRG 233
+DIWSLG ++E+ A + D L+ I LP S FR
Sbjct: 751 NYNGFAADIWSLGLTLFELYVGHFPFLQAGQRPDWATLMCAICFGDPPSLPETASPEFRD 930
Query: 234 LVKSMLRKNPELRPPASELLNHP 256
V+ L+K R ++LL HP
Sbjct: 931 FVECCLKKESGERWTTAQLLTHP 999
Score = 30.8 bits (68), Expect = 1.8
Identities = 16/48 (33%), Positives = 24/48 (49%)
Frame = +2
Query: 388 PSKISRSGSKHDTLPVSHAPSVKFPPPTRRASHPLPTRASMAKATLYT 435
P+ +S S S S +PS+ PPP + + PLP S + +L T
Sbjct: 101 PTSVSPSRSPPSAASASPSPSLPPPPPNQPPATPLPPPTSRSSPSLAT 244
>TC206365 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, partial (37%)
Length = 1866
Score = 111 bits (277), Expect = 1e-24
Identities = 80/268 (29%), Positives = 132/268 (48%), Gaps = 11/268 (4%)
Frame = +1
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTR---RSAHQEMELISKVR 59
Q++ + IG+GS+GS + LK++ L ++ + QE+ ++ ++
Sbjct: 433 QWQKGKLIGRGSYGSVYHATNLETGASCALKEVDLFPDDPKSADCIKQLEQEIRILRQLH 612
Query: 60 NPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDY 119
+P IV+Y S + G + I + Y G L + + G E + + +L L Y
Sbjct: 613 HPNIVQYYGSEIV-GDRLYIYMEYVHPGSLHKFMHEHCGA-MTESVVRNFTRHILSGLAY 786
Query: 120 LHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIP 179
LH +HRD+K +N+ + + ++L DFG++K+LT S+ G+P +M PEL+
Sbjct: 787 LHGTKTIHRDIKGANLLVDASGSVKLADFGVSKILTEKSYELSLKGSPYWMAPELMKAAI 966
Query: 180 YGSKS-------DIWSLGCCIYEMAALKPAFKAFD-IQALINKINKSIVAPLPTMYSSAF 231
S DIWSLGC I EM KP + F+ QA+ ++KS LP SS
Sbjct: 967 KKESSPDIAMAIDIWSLGCTIIEMLTGKPPWSEFEGPQAMFKVLHKS--PDLPESLSSEG 1140
Query: 232 RGLVKSMLRKNPELRPPASELLNHPHLQ 259
+ ++ R+NP RP A+ LL H +Q
Sbjct: 1141QDFLQQCFRRNPAERPSAAVLLTHAFVQ 1224
>TC228118 similar to UP|KP19_ARATH (Q39030) Serine/threonine-protein kinase
AtPK19 (Ribosomal-protein S6 kinase homolog) , partial
(65%)
Length = 1305
Score = 108 bits (270), Expect = 7e-24
Identities = 68/247 (27%), Positives = 126/247 (50%), Gaps = 1/247 (0%)
Frame = +1
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQ-EMELISKVR 59
++ +E+L+ +G+G+F VR K ++Y +K +R + ++ + E ++ +K+
Sbjct: 157 IDDFEILKVVGQGAFAKVYQVRKKDTSEIYAMKVMRKDKIMEKNHAEYMKAERDIWTKIE 336
Query: 60 NPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDY 119
+PF+V+ + S+ K + +++ + G L + F E+ + +++ A+ +
Sbjct: 337 HPFVVQLRYSFQTK-YRLYLVLDFVNGGHLFFQLYHQG--LFREDLARIYTAEIVSAVSH 507
Query: 120 LHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIP 179
LH N I+HRD+K NI L + + L DFGLAK ++S+ GT YM PE++
Sbjct: 508 LHANGIMHRDLKPENILLDADGHVMLTDFGLAKQFEESTRSNSMCGTLEYMAPEIILGKG 687
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
+ +D WS+G ++E KP F + + KI K + LP SS L+K +L
Sbjct: 688 HDKAADWWSVGVLLFETLTGKPPFCGGNRDKIQQKIVKDKI-KLPAFLSSEAHSLLKGLL 864
Query: 240 RKNPELR 246
+K R
Sbjct: 865 QKEQARR 885
>TC216364 similar to UP|Q9MAI7 (Q9MAI7) F12M16.4, partial (58%)
Length = 2224
Score = 107 bits (266), Expect = 2e-23
Identities = 70/204 (34%), Positives = 109/204 (53%), Gaps = 2/204 (0%)
Frame = +1
Query: 52 MELISKVRNPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLV 111
+ ++S+ R P+I EY S++ + + II+ Y G +AD I+ +G E + L
Sbjct: 1 ISVLSQCRCPYITEYYGSYLNQ-TKLWIIMEYMAGGSVADLIQ--SGPPLDEMSIACILR 171
Query: 112 QLLMALDYLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLT-SDDLASSIVGTPSYM 170
LL A+DYLH +HRD+K +NI L++N D+++ DFG++ LT + + VGTP +M
Sbjct: 172 DLLHAVDYLHSEGKIHRDIKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWM 351
Query: 171 CPELLADIP-YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSS 229
PE++ + Y K+DIWSLG EMA +P ++ I + L +S
Sbjct: 352 APEVIQNTDGYNEKADIWSLGITAIEMAKGEPPLADLHPMRVLFIIPRENPPQLDDHFSR 531
Query: 230 AFRGLVKSMLRKNPELRPPASELL 253
+ V L+K P RP A ELL
Sbjct: 532 PLKEFVSLCLKKVPAERPSAKELL 603
>TC227518 similar to UP|Q93VD3 (Q93VD3) At1g30270/F12P21_6 (CBL-interacting
protein kinase 23), partial (86%)
Length = 1492
Score = 106 bits (265), Expect = 3e-23
Identities = 69/260 (26%), Positives = 136/260 (51%), Gaps = 7/260 (2%)
Frame = +1
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKK---LYVLKKIRLARQSDRTRRSAHQEMELISKVR 59
+YE+ +G+G+F RH ++ + +L K +L + + +E+ + +R
Sbjct: 34 KYELGRTLGEGNFAKVKFARHVETRENVAIKILDKEKLLKH--KMIAQIKREISTMKLIR 207
Query: 60 NPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDY 119
+P ++ + K + I++ + G+L D I ++ + E++ K+ QL+ A+DY
Sbjct: 208 HPNVIRMYEVMASK-TKIYIVLEFVTGGELFDKIARSGRLK--EDEARKYFQQLICAVDY 378
Query: 120 LHVNHILHRDVKCSNIFLTKNQDIRLGDFGLA---KMLTSDDLASSIVGTPSYMCPELLA 176
H + HRD+K N+ L N +++ DFGL+ + + D L + GTP+Y+ PE++
Sbjct: 379 CHSRGVFHRDLKPENLLLDANGVLKVSDFGLSALPQQVREDGLLHTTCGTPNYVAPEVIN 558
Query: 177 DIPY-GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLV 235
+ Y G+K+D+WS G ++ + A F+ ++ AL KI K+ P +SS+ + L+
Sbjct: 559 NKGYDGAKADLWSCGVILFVLMAGYLPFEETNLSALYKKIFKAEFT-CPPWFSSSAKKLI 735
Query: 236 KSMLRKNPELRPPASELLNH 255
+L NP R +E++ +
Sbjct: 736 NKILDPNPATRITFAEVIEN 795
>TC227273 weakly similar to UP|KGS9_YEAST (P43637) Probable
serine/threonine-protein kinase YGL179C , partial (8%)
Length = 2373
Score = 106 bits (264), Expect = 3e-23
Identities = 61/223 (27%), Positives = 119/223 (53%), Gaps = 1/223 (0%)
Frame = +3
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPF 62
Q + ++G GSFG L R + + +K ++ R S R QE+ ++ K+R+
Sbjct: 1008 QLKYENKVGSGSFGD--LYRGTYCSQDVAIKVLKPERISTDMLREFAQEVYIMRKIRHKN 1181
Query: 63 IVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHV 122
+V++ + +CI+ + RG L D + K GV F L K + + ++YLH
Sbjct: 1182 VVQFIGACTRPPN-LCIVTEFMSRGSLYDFLHKQRGV-FKLPSLLKVAIDVSKGMNYLHQ 1355
Query: 123 NHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIPYGS 182
N+I+HRD+K +N+ + +N+ +++ DFG+A++ T + ++ GT +M PE++ PY
Sbjct: 1356 NNIIHRDLKTANLLMDENEVVKVADFGVARVQTQSGVMTAETGTYRWMAPEVIEHKPYDQ 1535
Query: 183 KSDIWSLGCCIYEMAALKPAFKAF-DIQALINKINKSIVAPLP 224
K+D++S G ++E+ + + +QA + + K + +P
Sbjct: 1536 KADVFSFGIALWELLTGELPYSCLTPLQAAVGVVQKGLRPTIP 1664
>TC205782 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, partial (91%)
Length = 1377
Score = 106 bits (264), Expect = 3e-23
Identities = 79/268 (29%), Positives = 132/268 (48%), Gaps = 10/268 (3%)
Frame = +2
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPF 62
+ E L +IG GS G+ V H+ ++Y LK I + RR H+E++++ V +
Sbjct: 278 ELERLNRIGSGSGGTVYKVVHRTSGRVYALKVI-YGHHEESVRRQIHREIQILRDVDDAN 454
Query: 63 IVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEK-LCKWLVQLLMALDYLH 121
+V+ + + ++ + +++ + + G L G + +E+ L Q+L L YLH
Sbjct: 455 VVKCHEMY-DQNSEIQVLLEFMDGGSL-------EGKHITQEQQLADLSRQILRGLAYLH 610
Query: 122 VNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELL-ADIP 179
HI+HRD+K SN+ + + +++ DFG+ ++L D +S VGT +YM PE + DI
Sbjct: 611 RRHIVHRDIKPSNLLINSRKQVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDIN 790
Query: 180 YGS----KSDIWSLGCCIYEMAALKPAF---KAFDIQALINKINKSIVAPLPTMYSSAFR 232
G DIWS G I E + F + D +L+ I S P S F+
Sbjct: 791 DGQYDAYAGDIWSFGVSILEFYMGRFPFAVGRQGDWASLMCAICMSQPPEAPPSASPHFK 970
Query: 233 GLVKSMLRKNPELRPPASELLNHPHLQP 260
+ L+++P R AS LL HP + P
Sbjct: 971 DFILRCLQRDPSRRWSASRLLEHPFIAP 1054
>CF922412
Length = 701
Score = 105 bits (263), Expect = 4e-23
Identities = 63/189 (33%), Positives = 101/189 (53%), Gaps = 1/189 (0%)
Frame = -2
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPF 62
+Y + ++IGKG++G +K++ L + QE++L+ + +
Sbjct: 565 KYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMQEIDLLKNLNHKN 386
Query: 63 IVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHV 122
IV+Y S K + I++ Y E G LA+ IK FPE + ++ Q+L L YLH
Sbjct: 385 IVKYLGSSKTKS-HLHIVLEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHE 209
Query: 123 NHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDL-ASSIVGTPSYMCPELLADIPYG 181
++HRD+K +NI TK ++L DFG+A LT D+ S+VGTP +M PE++
Sbjct: 208 QGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADVNTHSVVGTPYWMAPEVIEMAGVC 29
Query: 182 SKSDIWSLG 190
+ SDIWS+G
Sbjct: 28 AASDIWSVG 2
>TC216978 homologue to UP|Q8RXH5 (Q8RXH5) Osmotic stress-activated protein
kinase, partial (96%)
Length = 1626
Score = 104 bits (259), Expect = 1e-22
Identities = 81/264 (30%), Positives = 125/264 (46%), Gaps = 8/264 (3%)
Frame = +1
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M++YE+++ IG G+FG A L+RHK K+L +K I + D + +E+ +R+
Sbjct: 355 MDKYELVKDIGSGNFGVARLMRHKDTKELVAMKYIERGHKID---ENVAREIINHRSLRH 525
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P I+ +K+ V + I++ Y G+L + I A F E++ + QL+ + Y
Sbjct: 526 PNIIRFKEV-VLTPTHLGIVMEYAAGGELFERICSAG--RFSEDEARYFFQQLISGVSYC 696
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLG--DFGLAKMLTSDDLASSIVGTPSYMCPELLADI 178
H I HRD+K N L + RL DFG +K S VGTP+Y+ PE+L+
Sbjct: 697 HSMQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRR 876
Query: 179 PY-GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVA-----PLPTMYSSAFR 232
Y G +D+WS G +Y M F+ D K I+A P S R
Sbjct: 877 EYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQDCR 1056
Query: 233 GLVKSMLRKNPELRPPASELLNHP 256
L+ + NP R E+ +HP
Sbjct: 1057HLLSRIFVANPARRITIKEIKSHP 1128
>TC218712 similar to UP|Q40264 (Q40264) Protein kinase , partial (95%)
Length = 1223
Score = 104 bits (259), Expect = 1e-22
Identities = 80/269 (29%), Positives = 130/269 (47%), Gaps = 13/269 (4%)
Frame = +2
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELIS--KV 58
ME+YE+L+ IG G+F A LVR + +L+ +K I ++ D H + E+++ +
Sbjct: 50 MERYEILKDIGSGNFAVAKLVRDNYTNELFAVKFIERGQKIDE-----HVQREIMNHRSL 214
Query: 59 RNPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALD 118
++P I+ +K+ + + I++ Y G+L + I N F E++ + QL+ +
Sbjct: 215 KHPNIIRFKEVLLTP-THLAIVMEYASGGELFERI--CNAGRFSEDEARFFFQQLISGVS 385
Query: 119 YLHVNHILHRDVKCSNIFL--TKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLA 176
Y H I HRD+K N L + +++ DFG +K S VGTP+Y+ PE+L
Sbjct: 386 YCHSMQICHRDLKLENTLLDGSTAPRVKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLT 565
Query: 177 DIPYGSK-SDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMY-------- 227
Y K +D+WS G +Y M L A+ F+ A K+I L Y
Sbjct: 566 RKEYDGKIADVWSCGVTLYVM--LVGAY-PFEDPADPRNFKKTIGKILSVQYSVPDYVRV 736
Query: 228 SSAFRGLVKSMLRKNPELRPPASELLNHP 256
S R L+ + +PE R E+ NHP
Sbjct: 737 SMECRHLLSQIFVASPEKRITIPEIKNHP 823
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,996,957
Number of Sequences: 63676
Number of extensions: 443910
Number of successful extensions: 4006
Number of sequences better than 10.0: 696
Number of HSP's better than 10.0 without gapping: 3692
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3765
length of query: 601
length of database: 12,639,632
effective HSP length: 103
effective length of query: 498
effective length of database: 6,081,004
effective search space: 3028339992
effective search space used: 3028339992
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0322.12


