

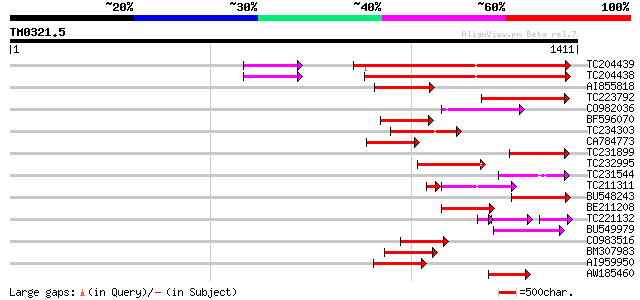
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0321.5
(1411 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 444 e-124
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 438 e-123
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 168 1e-41
TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polypro... 166 5e-41
CO982036 162 1e-39
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 154 3e-37
TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, parti... 148 2e-35
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 146 6e-35
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 144 3e-34
TC232995 142 1e-33
TC231544 weakly similar to UP|Q9FLR2 (Q9FLR2) Polyprotein-like, ... 131 2e-30
TC211311 weakly similar to UP|O24587 (O24587) Pol protein, parti... 101 1e-29
BU548243 126 6e-29
BE211208 125 1e-28
TC221132 weakly similar to UP|O23529 (O23529) RETROTRANSPOSON li... 81 3e-28
BU549979 124 3e-28
CO983516 120 3e-27
BM307983 119 7e-27
AI959950 116 6e-26
AW185460 114 2e-25
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 444 bits (1142), Expect = e-124
Identities = 232/555 (41%), Positives = 340/555 (60%), Gaps = 14/555 (2%)
Frame = +1
Query: 856 RPRTRSQNGIFRPNPRYALVHAQPTGILT-----------AFHTVTKPKGFKSAMKHP*W 904
+P RS I + +P+ ++ G+ T F + +PK K A+ W
Sbjct: 3061 QPDKRSSTRIQKMHPKELIIGDPNRGVTTRSREVEIVSNSCFVSKIEPKNVKEALTDEFW 3240
Query: 905 LAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQI 964
+ AM++EL +N W LVPRP TNV+G KW+F+ K + +G + R KARLVAQG+TQI
Sbjct: 3241 INAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQI 3420
Query: 965 PGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVD 1024
G D+ TF+PV + ++RL+L + ++L+Q+DVK+AFL+G+L E VY+EQP GF D
Sbjct: 3421 EGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFAD 3600
Query: 1025 PRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLV 1084
P P HV RL KALYGLKQAPRAW++RL+ FL + G+ D +LF + +
Sbjct: 3601 PTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQI 3780
Query: 1085 YVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAKYAHDL 1144
YVDDI+ G +L F+ ++ +EF + +G+L YFLGL++ D +FL Q++YA ++
Sbjct: 3781 YVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNI 3960
Query: 1145 LSRAMMLEASHVSTPLAAGSHL-VSSGEA--YFDPTHYRSLVGALQYLTITRPDLSYAVN 1201
+ + M ASH TP A +HL +S EA D + YRS++G+L YLT +RPD++YAV
Sbjct: 3961 VKKFGMENASHKRTP--APTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVG 4134
Query: 1202 TVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRST 1261
+++ P + H VKRIL+YV GT +G+ + S+P ++GY DADWA D R+ST
Sbjct: 4135 VCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKST 4314
Query: 1262 YGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPL 1321
G +LG+NL+SW +KKQ V+ S+ E+EY A ++ S+LVW+ +L E V
Sbjct: 4315 SGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVM-T 4491
Query: 1322 LLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALP 1381
L DN SA+ +++NPV H KHID+ H++ +LV + ++HV T Q+ADIFTKAL
Sbjct: 4492 LYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALD 4671
Query: 1382 RPLFEIFRSKLRVGL 1396
FE R KL + L
Sbjct: 4672 ANQFEKLRGKLGICL 4716
Score = 89.7 bits (221), Expect = 8e-18
Identities = 49/147 (33%), Positives = 75/147 (50%)
Frame = +1
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
IK +SD+G EF N++ S G+ H FS T QNG VE+K+R + E ML+
Sbjct: 2443 IKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAK 2622
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
+P W +A +TA YI NRV + + +++ P+ +FH FG +
Sbjct: 2623 ELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILADREQ 2802
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFN 727
K P+S IF GYS++ + ++ FN
Sbjct: 2803 RRKMDPKSDAGIFLGYSTNSRAYRVFN 2883
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 438 bits (1126), Expect = e-123
Identities = 225/516 (43%), Positives = 323/516 (61%), Gaps = 3/516 (0%)
Frame = +1
Query: 884 TAFHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKF 943
+ F + +PK K A+ W+ AM++EL +N W LVPRP TNV+G KW+F+ K
Sbjct: 3181 SCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKT 3360
Query: 944 HSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKN 1003
+ +G + R KARLVAQG+TQI G D+ TF+PV + ++RL+L + ++L+Q+DVK+
Sbjct: 3361 NEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKS 3540
Query: 1004 AFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSC 1063
AFL+G+L E Y+EQP GFVDP P HV RL KALYGLKQAPRAW++RL+ FL + G+
Sbjct: 3541 AFLNGYLNEEAYVEQPKGFVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRK 3720
Query: 1064 SRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLG 1123
D +LF + +YVDDI+ G +L F+ ++ +EF + +G+L YFLG
Sbjct: 3721 GGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLG 3900
Query: 1124 LEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHL-VSSGEA--YFDPTHYR 1180
L++ D +FL Q+KYA +++ + M ASH TP A +HL +S EA D + YR
Sbjct: 3901 LQVKQMEDSIFLSQSKYAKNIVKKFGMENASHKRTP--APTHLKLSKDEAGTSVDQSLYR 4074
Query: 1181 SLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSS 1240
S++G+L YLT +RPD++YAV +++ P + H VKRIL+YV GT +G+ + S
Sbjct: 4075 SMIGSLLYLTASRPDITYAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSD 4254
Query: 1241 PAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTAS 1300
++GY DADWA D R+ST G +LG NL+SW +KKQ V+ S+ E+EY A ++ S
Sbjct: 4255 SMLVGYCDADWAGSADDRKSTSGGCFYLGTNLISWFSKKQNCVSLSTAEAEYIAAGSSCS 4434
Query: 1301 ELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGR 1360
+LVW+ +L E V L DN SA+ +++NPV H KHID+ H++ +LV
Sbjct: 4435 QLVWMKQMLKEYNVEQDVM-TLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKV 4611
Query: 1361 LAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRVGL 1396
+ + HV T Q+ADIFTKAL FE R KL + L
Sbjct: 4612 ITLEHVDTEEQIADIFTKALDANQFEKLRGKLGICL 4719
Score = 92.4 bits (228), Expect = 1e-18
Identities = 51/147 (34%), Positives = 75/147 (50%)
Frame = +1
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
IK +SD+G EF N+K S G+ H FS T QNG VE+K+R + E ML+
Sbjct: 2446 IKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAK 2625
Query: 641 HVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYM 700
+P W +A +TA YI NRV + + +++ PT +FH FG +
Sbjct: 2626 ELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKHFHIFGSPCYILADREQ 2805
Query: 701 NNKFSPRSTPCIFFGYSSHHKGFKCFN 727
K P+S IF GYS++ + ++ FN
Sbjct: 2806 RRKMDPKSDAGIFLGYSTNSRAYRVFN 2886
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 168 bits (426), Expect = 1e-41
Identities = 78/151 (51%), Positives = 110/151 (72%)
Frame = -3
Query: 907 AMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPG 966
AM++EL+ +N W LV +P V+G KWVFR K G + R KARLVA+G+ Q G
Sbjct: 458 AMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEEG 279
Query: 967 FDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPR 1026
DY T++PV + +R++L++V + +++L+Q+DVK+AFL+G + E VY+EQPPGF P
Sbjct: 278 IDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIPD 99
Query: 1027 FPTHVCRLNKALYGLKQAPRAWFQRLSSFLL 1057
PTHV +L KALYGLKQAPRAW++R+S+FLL
Sbjct: 98 KPTHVYKLQKALYGLKQAPRAWYERISNFLL 6
>TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein, partial
(7%)
Length = 804
Score = 166 bits (421), Expect = 5e-41
Identities = 85/221 (38%), Positives = 138/221 (61%), Gaps = 3/221 (1%)
Frame = +1
Query: 1175 DPTHYRSLVGALQYLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLT 1234
D T +R L+G+L+YL +RP++ +AV+ +S+F++ P + H QA KR+LR + GT G+
Sbjct: 10 DVTEFRRLIGSLRYLCNSRPNICFAVSLISRFMKRPRLSHMQAAKRVLRLIKGTIGSGVL 189
Query: 1235 F---RRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESE 1291
F ++ P +LGY+D+DW R + +ST GY D ++ S+KKQ +A S+CE+E
Sbjct: 190 FPFKAKSGKPDLLGYTDSDWKRDPEQEKSTGGYLFMYNDAPVA*SSKKQDVIALSTCEAE 369
Query: 1292 YRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHF 1351
Y A + A + VW++NLL EL++R LL DN+SA+ +A++P H +KHI+L H+
Sbjct: 370 YVAASLGACQAVWMMNLLEELKLRERKPVNLLIDNKSAINLAKHPTLHGRSKHIELRFHY 549
Query: 1352 VCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSKL 1392
+ + V+ G + V + QLAD+ TK + F+ S+L
Sbjct: 550 IRDQVSKGNVTVEYCKAEEQLADLMTKPIQVSRFKQICSEL 672
>CO982036
Length = 674
Score = 162 bits (409), Expect = 1e-39
Identities = 95/212 (44%), Positives = 128/212 (59%), Gaps = 5/212 (2%)
Frame = -2
Query: 1074 YKGHT-TLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDG 1132
YK H T+YLLVYVD II+TGS +L+ ++LN+ F +K LGKL YF+ +E+ PD
Sbjct: 673 YKTHILTVYLLVYVD-IIITGSSCTLIQNLTSKLNSSFPLKLLGKLDYFVEIEVKSMPDL 497
Query: 1133 LFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSSGEAYFD-PTHYRSLVGALQYLTI 1191
LF + ++ R +A +S+P+ L S F PT YRS+VGALQY T+
Sbjct: 496 LFSLRTSI-FEIFCRKPR*QAQPISSPMTTTCKLSKSDSDLFSGPTFYRSVVGALQYTTV 320
Query: 1192 TRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVL---GYSD 1248
RP++S+AVN V QF+ P H+ VKRILRY+ G+ +GL + S L G+ D
Sbjct: 319 IRPEISFAVNKVCQFMSNPLDSHWTEVKRILRYLKGSLSYGL*LKPAISSQPLPIRGFCD 140
Query: 1249 ADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQ 1280
ADWA D +RST G A+FLG NL+SW KQ
Sbjct: 139 ADWASAVDDKRSTSGAAVFLGPNLISWWXXKQ 44
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 154 bits (388), Expect = 3e-37
Identities = 75/130 (57%), Positives = 91/130 (69%)
Frame = -2
Query: 924 VPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVR 983
VP P VG +WV+ K G V+RLKARLVA+G+TQ+ G DY TFSPV K TTVR
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 984 LILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQ 1043
L L+ + W LHQLD+KNAFLHG L E +YMEQPPGFV VC+L+++LYGLKQ
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 1044 APRAWFQRLS 1053
+PRAWF + S
Sbjct: 46 SPRAWFGKFS 17
>TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, partial (10%)
Length = 558
Score = 148 bits (373), Expect = 2e-35
Identities = 86/180 (47%), Positives = 112/180 (61%), Gaps = 2/180 (1%)
Frame = +1
Query: 947 GTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFL 1006
GT+++ KARLVA+ +TQ+ G DY+ TFSPV K V L+ S V+ W L LD KNAFL
Sbjct: 28 GTIDQFKARLVAKSYTQVYGQDYTGTFSPVAKMAYVHLLWSMAVVCHWPLF*LDAKNAFL 207
Query: 1007 HGHLTETVYMEQPPGFV-DPRFPTHVCRLNKALYGLKQAPRAW-FQRLSSFLLRHGFSCS 1064
HG+L E VYMEQP GFV VC+L ++ YGLKQ+PRAW F + + +
Sbjct: 208 HGYLEEEVYMEQPLGFVAQGESSNMVCQLCRSFYGLKQSPRAWPFLYCGAAI---WYDSH 378
Query: 1065 RADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGL 1124
AD S+F+ + +YL+VYVDDI +TGSD +T L +F K LGKL YFLG+
Sbjct: 379 EADHSVFYCHSPQGCIYLIVYVDDIGITGSDQHGIT*LK*XLCCQFQTKDLGKLRYFLGI 558
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 146 bits (369), Expect = 6e-35
Identities = 70/132 (53%), Positives = 87/132 (65%)
Frame = +3
Query: 888 TVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDG 947
++T P + A+ HP W AM DE+ AL N TW LVP P VG +WV+ K +G
Sbjct: 9 SLTVPSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGPNG 188
Query: 948 TVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLH 1007
V+RLKARLVA+G+TQ+ G +Y TFSPV TTVRL L+ + W LHQLD+KNAFLH
Sbjct: 189 KVDRLKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAFLH 368
Query: 1008 GHLTETVYMEQP 1019
G L E +YMEQP
Sbjct: 369 GDLEEDIYMEQP 404
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment), partial
(30%)
Length = 687
Score = 144 bits (363), Expect = 3e-34
Identities = 72/148 (48%), Positives = 93/148 (62%)
Frame = +2
Query: 1245 GYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVW 1304
GY DADWA C RRST GY +F+G NL+SW +KKQ VARSS E+EYR+MA EL+W
Sbjct: 23 GYCDADWAGCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMAMVTCELMW 202
Query: 1305 LLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVR 1364
+ L ELR L DNQ+AL +A NPV H+ KHI++DCHF+ E + S +
Sbjct: 203 IKQFLQELRFCEELQMKLYCDNQAALHIASNPVFHERTKHIEIDCHFIREKLLSKEIVTE 382
Query: 1365 HVPTSLQLADIFTKALPRPLFEIFRSKL 1392
+ ++ Q DI TK+L P +I SKL
Sbjct: 383 FIGSNDQPVDILTKSLRGPKIQIVCSKL 466
>TC232995
Length = 1009
Score = 142 bits (357), Expect = 1e-33
Identities = 76/171 (44%), Positives = 107/171 (62%), Gaps = 2/171 (1%)
Frame = +2
Query: 1016 MEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYK 1075
+EQPPGF P HV +L KALYGLKQAPRAW++RLS+FLL FS + D +LF K
Sbjct: 2 VEQPPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRK 181
Query: 1076 GHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDGLFL 1135
+ L + +YVDDII ++ SL +F + +EF + +G+L YFLGL+I T G+F+
Sbjct: 182 HNDILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQIKQTQ*GIFI 361
Query: 1136 GQAKYAHDLLSRAMMLEASHVSTPLAAGSHL--VSSGEAYFDPTHYRSLVG 1184
Q+KY +L+ R M A H+STP++ +L SG++ D YR +G
Sbjct: 362 NQSKYCKELIKRFGMDSAKHMSTPMSTNCYLDKDESGQS-IDIKQYRDAIG 511
>TC231544 weakly similar to UP|Q9FLR2 (Q9FLR2) Polyprotein-like, partial (16%)
Length = 662
Score = 131 bits (329), Expect = 2e-30
Identities = 82/179 (45%), Positives = 105/179 (57%), Gaps = 3/179 (1%)
Frame = +3
Query: 1217 AVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWS 1276
A R+L+Y+ G GL+F R S +LG+SDADWA C D+ +S Y FLG +L+SW
Sbjct: 18 AATRVLKYLKGCPRKGLSFSRESPIQILGFSDADWATCIDSSKSITWYCFFLGSSLISWK 197
Query: 1277 AKKQPTVARSSCESE--YRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQ 1334
AKKQ TV+RSS SE YRA+ +T EL WL LL +L V L+ DNQSAL
Sbjct: 198 AKKQNTVSRSSSSSEAKYRALTSTTCELQWLTYLLKDLHV-----TLIYCDNQSAL--Q* 356
Query: 1335 NPVAHKHAKHIDLDCHFVCELVASGRL-AVRHVPTSLQLADIFTKALPRPLFEIFRSKL 1392
P+ + +++DCH V E G + + V +S QLADIFTKAL LF SKL
Sbjct: 357 LPIKVIYHGQLEIDCHIVREKTQQGLMHCLLPVSSSNQLADIFTKALSPKLFSSNLSKL 533
>TC211311 weakly similar to UP|O24587 (O24587) Pol protein, partial (15%)
Length = 1213
Score = 101 bits (252), Expect(2) = 1e-29
Identities = 64/189 (33%), Positives = 97/189 (50%), Gaps = 2/189 (1%)
Frame = +3
Query: 1075 KGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDGLF 1134
K T L + +YVDDII + + +F + F G+L + LGL+I G+F
Sbjct: 474 KKETFLIIHIYVDDIIFGATSKRMCKEFFELMKDGFETSMKGELKFLLGLQIIQKVYGIF 653
Query: 1135 LGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSSGEA--YFDPTHYRSLVGALQYLTIT 1192
+ Q KY L R M EA ++TP+ S ++ E + Y ++ +L YLT +
Sbjct: 654 IHQEKYTKSHLKRFRMDEAKPMATPMHR-STIIDKDEKGNHTS*KEYSGMIDSLSYLTSS 830
Query: 1193 RPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWA 1252
RPD+ + V ++F P + H AVKRILRY+ GT + L F++ S +LGY D +A
Sbjct: 831 RPDIVFVVCLCARFQSYPKISHVTAVKRILRYLVGTTNHCLWFKKRSEFDLLGYCDVYFA 1010
Query: 1253 RCTDTRRST 1261
R+ST
Sbjct: 1011 GDKVERKST 1037
Score = 48.1 bits (113), Expect(2) = 1e-29
Identities = 21/34 (61%), Positives = 28/34 (81%)
Frame = +2
Query: 1038 LYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLF 1071
+YGLKQA RAW++RLSSFL+ +GF+ DP+LF
Sbjct: 362 VYGLKQALRAWYERLSSFLVSNGFTRGITDPALF 463
>BU548243
Length = 599
Score = 126 bits (317), Expect = 6e-29
Identities = 68/147 (46%), Positives = 93/147 (63%)
Frame = -1
Query: 1248 DADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLN 1307
DA WA D RST G AIFLG NL+SW ++KQ A+SS E+EYR++A T++EL W+
Sbjct: 587 DAGWASDVDDHRSTLGSAIFLGPNLISWWSRKQQVTAQSSTEAEYRSIAQTSAELTWIQA 408
Query: 1308 LLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVP 1367
LL EL++ + P++L DN+SA+ +A N V H KH+++D FV E V S +L + H+P
Sbjct: 407 LLMELQIPFT-PPVILCDNKSAVAIAHNLVFHSRTKHMEIDVFFVHEKVLSKQLQIFHIP 231
Query: 1368 TSLQLADIFTKALPRPLFEIFRSKLRV 1394
Q A I TK L F +SKL V
Sbjct: 230 ALDQWAGILTKPLSSARFTFLKSKLTV 150
>BE211208
Length = 413
Score = 125 bits (315), Expect = 1e-28
Identities = 66/134 (49%), Positives = 89/134 (66%), Gaps = 2/134 (1%)
Frame = +2
Query: 1075 KGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDG-L 1133
K +YLLVYVDDII+TG L+ + LN+ F++K LG+L YFLG+E+ +TP G +
Sbjct: 11 KDRNLVYLLVYVDDIIITGRSNYLIQSLVHHLNSNFSLKQLGQLDYFLGIEVHHTPTGSV 190
Query: 1134 FLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSSGEAYF-DPTHYRSLVGALQYLTIT 1192
L Q+KY DLL + M EA +S+P+ L +G+ DPT YRS+VGALQY TIT
Sbjct: 191 LLTQSKYICDLLHKTDMAEAKPISSPMVTNLRLSKNGDDLLSDPTMYRSVVGALQYPTIT 370
Query: 1193 RPDLSYAVNTVSQF 1206
RP++S+A N V QF
Sbjct: 371 RPEISFAANKVCQF 412
>TC221132 weakly similar to UP|O23529 (O23529) RETROTRANSPOSON like protein,
partial (5%)
Length = 799
Score = 81.3 bits (199), Expect(3) = 3e-28
Identities = 40/104 (38%), Positives = 61/104 (58%)
Frame = +1
Query: 1198 YAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDT 1257
++++T SQF++ PT QA KR+LRY+ GT FGL R + + + DA+W T
Sbjct: 115 HSLSTSSQFMKDPTKIRMQATKRVLRYLKGTIDFGLQLRSSPDQHLRAFYDANWVDNTSD 294
Query: 1258 RRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASE 1301
RST Y ++ G +++SWS KKQ + +SS + EY + T E
Sbjct: 295 IRSTGAYVVYFGLSVISWSCKKQSIIDKSSTKVEYHKITTTIIE 426
Score = 53.1 bits (126), Expect(3) = 3e-28
Identities = 30/82 (36%), Positives = 45/82 (54%)
Frame = +2
Query: 1319 TPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTK 1378
+P + S N A+++ NPV H KH+ +D FV +LVA+ +L V HVP+ D+FTK
Sbjct: 446 SPTMYSYNIGAMYLCANPVFHLCMKHLTIDHLFVQDLVANKQLRVSHVPSCH*HVDLFTK 625
Query: 1379 ALPRPLFEIFRSKLRVGLNPTL 1400
AL + K+ V T+
Sbjct: 626 ALVSSRHKFMMDKIGVVSTTTI 691
Score = 31.2 bits (69), Expect(3) = 3e-28
Identities = 15/40 (37%), Positives = 24/40 (59%)
Frame = +3
Query: 1165 HLVSSGEAYFDPTHYRSLVGALQYLTITRPDLSYAVNTVS 1204
+L S D Y LV +LQYL++T PD+++ +N +S
Sbjct: 18 NLASGDVPSCDGIVYCQLVDSLQYLSLTCPDIAFPINKLS 137
>BU549979
Length = 615
Score = 124 bits (311), Expect = 3e-28
Identities = 63/180 (35%), Positives = 105/180 (58%), Gaps = 2/180 (1%)
Frame = -1
Query: 1203 VSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTY 1262
+ ++ P ++H++ K+++RY+ GT+ + L +++T+ V+GYSD+D+A C D+RRST
Sbjct: 612 LGRYQSNPGIDHWKTAKKVMRYLQGTKDYMLMYKQTNCLEVIGYSDSDFAGCVDSRRSTS 433
Query: 1263 GYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRV--RLSATP 1320
GY L D ++SW + KQ +A S+ E E+ S VWL + + LRV +S
Sbjct: 432 GYIFMLADGVVSWRSSKQTLIATSTMEVEFVPCFEATSHGVWLKSFMSSLRVVDSISRPL 253
Query: 1321 LLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKAL 1380
L DN +A+FMA+N + +KHID+ + E V ++ + HV T L + D TK +
Sbjct: 252 KLYCDNFAAVFMAKNNKSGNRSKHIDIKYLVIRERVKEKKVVIEHVNTELMIVDPLTKGM 73
>CO983516
Length = 724
Score = 120 bits (302), Expect = 3e-27
Identities = 56/118 (47%), Positives = 79/118 (66%)
Frame = +2
Query: 973 FSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVC 1032
F PV + ++RL+L + ++L+Q+DVK+AFL+G+L E VY+EQP GF+DP P HV
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 1033 RLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDII 1090
RL KALYGLKQAPRAW++RL+ L + G+ D +LF + +YVDDI+
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIV 718
>BM307983
Length = 406
Score = 119 bits (299), Expect = 7e-27
Identities = 63/134 (47%), Positives = 82/134 (61%), Gaps = 1/134 (0%)
Frame = +2
Query: 933 VGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVK-ATTVRLILSHVVL 991
VG +W++ K+ +D T++R KARLVA+G+ Q G DY TF+ K +
Sbjct: 2 VGCRWIYTVKY*ADDTLDRYKARLVAKGYIQTYGIDYEETFAQWQK*IQSGSSSP*QQAQ 181
Query: 992 NDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQR 1051
W++HQ DVKNAFLHG L E VYME PPG+ VCRL KALYGLKQ+PRAWF R
Sbjct: 182 FGWEMHQFDVKNAFLHGSLEEEVYMEIPPGYGASNGGNKVCRLKKALYGLKQSPRAWFGR 361
Query: 1052 LSSFLLRHGFSCSR 1065
+ +L G+ S+
Sbjct: 362 FTQAMLSLGYKQSQ 403
>AI959950
Length = 466
Score = 116 bits (291), Expect = 6e-26
Identities = 58/132 (43%), Positives = 83/132 (61%)
Frame = -1
Query: 905 LAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQI 964
+ AM++EL KN LV P VVG+KW+F K DG V R KARLVA+G++Q
Sbjct: 397 MKAMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQ 218
Query: 965 PGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVD 1024
G DY TF+ V + + ++LS ++ +L+Q+DVK+AFL+G + + VY+EQPPGF +
Sbjct: 217 EGIDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGFEN 38
Query: 1025 PRFPTHVCRLNK 1036
HV +LNK
Sbjct: 37 ETLHQHVFKLNK 2
>AW185460
Length = 411
Score = 114 bits (286), Expect = 2e-25
Identities = 53/105 (50%), Positives = 77/105 (72%)
Frame = +2
Query: 1192 TRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADW 1251
TRPD+ YA + +S+F+Q+P+ HF A KRILRY+ GT+ FG+ + ++ +LGY+D+DW
Sbjct: 92 TRPDIMYATSLLSRFMQSPSQIHFGAGKRILRYLQGTKAFGIWYTTETNSELLGYTDSDW 271
Query: 1252 ARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMA 1296
A TD +ST GYA LG + SW++KKQ TVA+S+ E+EY A+A
Sbjct: 272 AGSTDDMKSTSGYAFSLGSGMFSWASKKQATVAQSTAEAEYVAVA 406
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.347 0.151 0.556
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 84,056,231
Number of Sequences: 63676
Number of extensions: 1747877
Number of successful extensions: 50673
Number of sequences better than 10.0: 1570
Number of HSP's better than 10.0 without gapping: 19607
Number of HSP's successfully gapped in prelim test: 769
Number of HSP's that attempted gapping in prelim test: 4249
Number of HSP's gapped (non-prelim): 31166
length of query: 1411
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1302
effective length of database: 5,698,948
effective search space: 7420030296
effective search space used: 7420030296
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0321.5


