

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0321.10
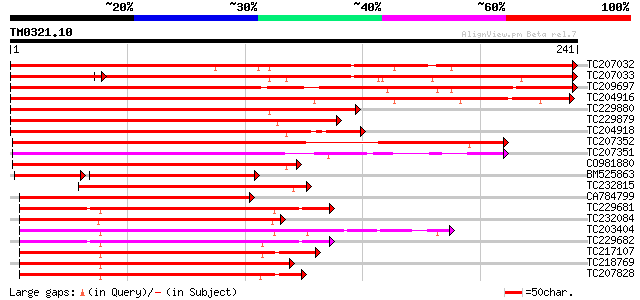
(241 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207032 similar to UP|Q8H0H0 (Q8H0H0) Myb-like protein, partial... 337 4e-93
TC207033 similar to UP|Q941B3 (Q941B3) AT5g67300/K8K14_2, partia... 256 1e-84
TC209697 similar to UP|Q8H0H0 (Q8H0H0) Myb-like protein, partial... 296 7e-81
TC204916 similar to UP|Q8H0H0 (Q8H0H0) Myb-like protein, partial... 269 7e-73
TC229880 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 229 9e-61
TC229879 homologue to UP|Q8H0H0 (Q8H0H0) Myb-like protein, parti... 225 1e-59
TC204918 similar to UP|O23160 (O23160) Myb-related protein (MYB ... 217 4e-57
TC207352 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucros... 184 3e-47
TC207351 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucros... 181 3e-46
CO981880 176 9e-45
BM525863 126 5e-37
TC232815 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucros... 142 1e-34
CA784799 119 1e-27
TC229681 UP|Q9S7E3 (Q9S7E3) GmMYB29A1 protein, complete 117 5e-27
TC232084 UP|Q84PP2 (Q84PP2) Transcription factor MYB101 (Fragmen... 115 2e-26
TC203404 GmMYB16 115 2e-26
TC229682 UP|Q9XIU9 (Q9XIU9) GmMYB29A2 protein, complete 115 2e-26
TC217107 homologue to UP|Q7X9I1 (Q7X9I1) MYB transcription facto... 112 1e-25
TC218769 UP|Q9XIU8 (Q9XIU8) GmMYB29A2 protein, complete 112 1e-25
TC207828 homologue to UP|Q6SL45 (Q6SL45) MYB6, partial (52%) 112 2e-25
>TC207032 similar to UP|Q8H0H0 (Q8H0H0) Myb-like protein, partial (43%)
Length = 1001
Score = 337 bits (863), Expect = 4e-93
Identities = 183/272 (67%), Positives = 203/272 (74%), Gaps = 31/272 (11%)
Frame = +3
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
MDRVKGPWSPEEDEAL RLVQ HGP+NW+ ISKS+PGRSGKSCRLRWCNQLSP+V HRPF
Sbjct: 90 MDRVKGPWSPEEDEALRRLVQAHGPRNWSVISKSVPGRSGKSCRLRWCNQLSPQVAHRPF 269
Query: 61 TPEEDSAIIRAHARFGNKWATIARIL-NGRTDNAVKNHWNSTLKR-KCSAV-DTAVDHRP 117
+P+ED AI+RAHARFGNKWATIAR+L NGRTDNAVKNHWNSTLKR KCSAV D D P
Sbjct: 270 SPDEDEAIVRAHARFGNKWATIARLLNNGRTDNAVKNHWNSTLKRKKCSAVSDDVTDRPP 449
Query: 118 LKRSASVGAGCLNPSSPTGSAMSDPGPPAAISTDAPIAVPAAVVV----PATDPATSLSL 173
LKRSASVG LNP SP+GS +SDP P A+S +P A ++ A DPATSLSL
Sbjct: 450 LKRSASVGPAHLNPGSPSGSDLSDPSLP-ALSNPSPSAHYRPNLMETASSACDPATSLSL 626
Query: 174 SLPGFDSSGSGSG------------------------SKQLFGAEFLAVMQEMIRKEVRS 209
SLPGF GSGS KQ+F AEFL VMQEMIRKEVRS
Sbjct: 627 SLPGF---GSGSNPVCAPRPVSVSPPSPPPAEPVEQRGKQMFNAEFLRVMQEMIRKEVRS 797
Query: 210 YMSGMEEKNGACMPAEAIGNAVMKRMGISNVK 241
YMSGME+KNG +P EAIGNAV+KRMGISN++
Sbjct: 798 YMSGMEQKNGFRIPTEAIGNAVVKRMGISNIE 893
>TC207033 similar to UP|Q941B3 (Q941B3) AT5g67300/K8K14_2, partial (42%)
Length = 1164
Score = 256 bits (655), Expect(2) = 1e-84
Identities = 148/228 (64%), Positives = 164/228 (71%), Gaps = 23/228 (10%)
Frame = +2
Query: 37 GRSGKSCRLRWCNQLSPEVEHRPFTPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKN 96
G SGKSCRLRWCNQLSP+V HRPF+ EED AII AHA+FGNKWATIAR+LNGRTDNAVKN
Sbjct: 227 GDSGKSCRLRWCNQLSPQVAHRPFSQEEDEAIIMAHAKFGNKWATIARLLNGRTDNAVKN 406
Query: 97 HWNSTLKRKCSAV--DTAVDHR-PLKRSASVGAGCLNPSSPTGSAMSDPGPPAAISTDAP 153
HWNSTLKRK SAV D V HR PLKRS SVG LNP+SP+ S +SDPG P A+S +P
Sbjct: 407 HWNSTLKRKSSAVSDDDVVTHRQPLKRSNSVGPAHLNPASPSVSDLSDPGLP-ALSNPSP 583
Query: 154 IA--VP--AAVVVPATDPATSLSLSLPGF---------------DSSGSGSGSKQLFGAE 194
A +P A DPATSLSLSLPGF + KQ+F AE
Sbjct: 584 SAHYMPNLMETASSAPDPATSLSLSLPGFTVPQPVSPPPPPLPLPAETVEERGKQMFNAE 763
Query: 195 FLAVMQEMIRKEVRSYMSGMEE-KNGACMPAEAIGNAVMKRMGISNVK 241
FL VMQEMI+KEVRSYMSGMEE KNG M EAIGNAV+KRMGISN++
Sbjct: 764 FLRVMQEMIKKEVRSYMSGMEEQKNGFRMQTEAIGNAVVKRMGISNIE 907
Score = 74.3 bits (181), Expect(2) = 1e-84
Identities = 33/41 (80%), Positives = 35/41 (84%)
Frame = +1
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGK 41
MDRVKGPWSPEEDEAL LVQ HGP+NW+ ISKSIPGR K
Sbjct: 118 MDRVKGPWSPEEDEALRALVQAHGPRNWSVISKSIPGRFRK 240
>TC209697 similar to UP|Q8H0H0 (Q8H0H0) Myb-like protein, partial (51%)
Length = 882
Score = 296 bits (757), Expect = 7e-81
Identities = 159/268 (59%), Positives = 186/268 (69%), Gaps = 27/268 (10%)
Frame = +1
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
MDR+KGPWSPEEDEAL RLVQ +GP+NW+ ISKSIPGRSGKSCRLRWCNQLSPEVE RPF
Sbjct: 28 MDRIKGPWSPEEDEALRRLVQTYGPRNWSVISKSIPGRSGKSCRLRWCNQLSPEVERRPF 207
Query: 61 TPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVDHRPLKR 120
T EED AI++AHARFGNKWATIAR LNGRTDNA+KNHWNSTLKRKCS + + RPLKR
Sbjct: 208 TAEEDEAILKAHARFGNKWATIARFLNGRTDNAIKNHWNSTLKRKCS--EPLSEPRPLKR 381
Query: 121 SASVGAGCLNPSSPTGSAMSDPGPPAAISTDAPIAVPAA------VVVPATDPATSLSLS 174
SA+V S +GS +SD G P ++ + V + TDPAT LSLS
Sbjct: 382 SATVS------GSQSGSDLSDSGLPIILARSVSVTVAPSNHLAETASSSVTDPATLLSLS 543
Query: 175 LPGFDS-SGSGSG--------------------SKQLFGAEFLAVMQEMIRKEVRSYMSG 213
LPGFDS G+ +G KQLF EF+ VMQEMIR EVR+YMS
Sbjct: 544 LPGFDSCDGANNGPGPNQGPSCGPFQEIPMLGSQKQLFSQEFMKVMQEMIRVEVRNYMSV 723
Query: 214 MEEKNGACMPAEAIGNAVMKRMGISNVK 241
+ E+NG CM +AI N+V++RMGI V+
Sbjct: 724 L-ERNGVCMQTDAIRNSVLERMGIGRVE 804
>TC204916 similar to UP|Q8H0H0 (Q8H0H0) Myb-like protein, partial (51%)
Length = 1505
Score = 269 bits (688), Expect = 7e-73
Identities = 154/296 (52%), Positives = 179/296 (60%), Gaps = 56/296 (18%)
Frame = +2
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
MDR+KGPWSPEEDEAL +LV+ HGP+NW+ ISKSIPGRSGKSCRLRWCNQLSP+VEHR F
Sbjct: 80 MDRIKGPWSPEEDEALQKLVEKHGPRNWSLISKSIPGRSGKSCRLRWCNQLSPQVEHRAF 259
Query: 61 TPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVDHRPLKR 120
T EED IIRAHARFGNKWATIAR+L+GRTDNA+KNHWNSTLKRKCS+ + +PLKR
Sbjct: 260 TAEEDDTIIRAHARFGNKWATIARLLHGRTDNAIKNHWNSTLKRKCSSTMIDDNTQPLKR 439
Query: 121 SASVGAGC----------LNPSSPTGSAMSDPGPPAAISTDAPIAVPAAVVV-------- 162
S S GA P SP+GS +S+ P A S+ VP V V
Sbjct: 440 SVSAGAAIPVSTGLYMNPPTPGSPSGSDVSESSLPVASSSHVFRPVPRTVKVLPPVETTS 619
Query: 163 PATDPATSLSLSLPGFDSSGSGSGSKQL-------------------------------- 190
+ DP TSLSLSLPG S S +
Sbjct: 620 SSNDPPTSLSLSLPGVGVVDSSEVSNRTTEPIHFTPPMPHRSNTMPLLPMMTSVSATVMA 799
Query: 191 ---FGAEFLAVMQEMIRKEVRSYMSGMEEKNGACMPA---EAIGNAVMKRMGISNV 240
F AEF +VMQEMIRKEVRSY+ + +NG C A + NA +KR+GIS V
Sbjct: 800 PFNFSAEFRSVMQEMIRKEVRSYIE-LHNQNGMCFQAAVNDGFRNASVKRIGISRV 964
>TC229880 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose-responsive
element binding factor, partial (35%)
Length = 583
Score = 229 bits (584), Expect = 9e-61
Identities = 107/155 (69%), Positives = 124/155 (79%), Gaps = 6/155 (3%)
Frame = +1
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
MDR+KGPW+P+EDEAL +LV+ HGP+NW+ IS+SIPGRSGKSCRLRWCNQLSP+VEHR F
Sbjct: 100 MDRIKGPWNPKEDEALQKLVERHGPRNWSLISRSIPGRSGKSCRLRWCNQLSPQVEHRAF 279
Query: 61 TPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAV------DTAVD 114
TPEED IIRAHARFGNKWATIAR+L+GRTDNA+KNHWNSTLKRKC++ AV
Sbjct: 280 TPEEDETIIRAHARFGNKWATIARLLSGRTDNAIKNHWNSTLKRKCASFMMAGDEAVAVS 459
Query: 115 HRPLKRSASVGAGCLNPSSPTGSAMSDPGPPAAIS 149
RPLKRS S GA P SP+GS S+ P +S
Sbjct: 460 PRPLKRSFSAGAAVPPPGSPSGSDFSESSAPGVVS 564
>TC229879 homologue to UP|Q8H0H0 (Q8H0H0) Myb-like protein, partial (38%)
Length = 681
Score = 225 bits (574), Expect = 1e-59
Identities = 106/148 (71%), Positives = 121/148 (81%), Gaps = 7/148 (4%)
Frame = +1
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
MDR+KGPWSPEEDEAL +LV+ HGP+NW+ IS+SIPGRSGKSCRLRWCNQLSP+VEHR F
Sbjct: 136 MDRIKGPWSPEEDEALQKLVERHGPRNWSLISRSIPGRSGKSCRLRWCNQLSPQVEHRAF 315
Query: 61 TPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTA-------V 113
TPEED IIRAHARFGNKWATIAR+L+GRTDNA+KNHWNSTLKRKC++ TA
Sbjct: 316 TPEEDETIIRAHARFGNKWATIARLLSGRTDNAIKNHWNSTLKRKCASFMTADEAAAGGF 495
Query: 114 DHRPLKRSASVGAGCLNPSSPTGSAMSD 141
RPLKRS S G P SP+GS +S+
Sbjct: 496 SPRPLKRSFSAGTAVPPPGSPSGSDVSE 579
Score = 28.9 bits (63), Expect = 2.2
Identities = 26/78 (33%), Positives = 34/78 (43%), Gaps = 13/78 (16%)
Frame = +2
Query: 117 PLKRSASVGAGCLNPSS-PTGSAMSDPGPPAAISTDAPIAVPAAVVV------------P 163
P+ S G LN S+ P+ M P +A + + PA + P
Sbjct: 404 PITPSRITGTQRLNASARPS*PPMKPPPAASARGRSSDRSAPARRCLLQEAPQDPT*ASP 583
Query: 164 ATDPATSLSLSLPGFDSS 181
A PA SLSLSLPG +SS
Sbjct: 584 ALLPAWSLSLSLPGVESS 637
>TC204918 similar to UP|O23160 (O23160) Myb-related protein (MYB
transcription factor), partial (41%)
Length = 565
Score = 217 bits (552), Expect = 4e-57
Identities = 107/152 (70%), Positives = 122/152 (79%), Gaps = 1/152 (0%)
Frame = +3
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
MDR+KGPWSPEEDEAL +LV+ HGP+NW+ ISKSIPGRSGKSCRLRWCNQLSP+VEHR F
Sbjct: 93 MDRIKGPWSPEEDEALQKLVEKHGPRNWSLISKSIPGRSGKSCRLRWCNQLSPQVEHRAF 272
Query: 61 TPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVDHR-PLK 119
T EED IIRAHARFGNKWATIAR+L+GRTDNA+KNHWNSTLKRKC++ D+ PLK
Sbjct: 273 THEEDDTIIRAHARFGNKWATIARLLHGRTDNAIKNHWNSTLKRKCTSTMIDDDNTPPLK 452
Query: 120 RSASVGAGCLNPSSPTGSAMSDPGPPAAISTD 151
RS S GA P S TG M+ P P + +D
Sbjct: 453 RSLSAGAAI--PVS-TGLYMNPPTPGSPSGSD 539
>TC207352 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose-responsive
element binding factor, partial (58%)
Length = 811
Score = 184 bits (467), Expect = 3e-47
Identities = 99/214 (46%), Positives = 129/214 (60%), Gaps = 3/214 (1%)
Frame = +1
Query: 2 DRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFT 61
DR+KGPWS EED LT LV+ +GP+NW+ IS+ I GRSGKSCRLRWCNQLSP VEHRPF+
Sbjct: 124 DRIKGPWSAEEDRILTGLVERYGPRNWSLISRYIKGRSGKSCRLRWCNQLSPAVEHRPFS 303
Query: 62 PEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVDHRPLKRS 121
+ED II AHA++GN+WATIAR+L GRTDNAVKNHWNSTLKR+ ++ V+H + +
Sbjct: 304 AQEDDTIIAAHAQYGNRWATIARLLPGRTDNAVKNHWNSTLKRRAKGININVNHANNEVA 483
Query: 122 ASVGAGCLNPSSPTGSAMSDPGPPAAISTDAPIAVPAAVVVPATDPATSLSLSLPGFDSS 181
AS A P+ DP T+L+L+ PG +
Sbjct: 484 ASSSA------------------------------PSRHFDFEDDPLTALTLAPPGIANG 573
Query: 182 GSGSGSKQLFGAE---FLAVMQEMIRKEVRSYMS 212
+ A F +M+++I +EVR Y+S
Sbjct: 574 TVAEDAVPDHRASPEGFWDMMRDVIAREVREYVS 675
>TC207351 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose-responsive
element binding factor, partial (52%)
Length = 974
Score = 181 bits (458), Expect = 3e-46
Identities = 100/212 (47%), Positives = 127/212 (59%), Gaps = 1/212 (0%)
Frame = +3
Query: 2 DRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFT 61
DR+KGPWS +ED LTRLV+ +GP+NW+ IS+ I GRSGKSCRLRWCNQLSP VEHRPF+
Sbjct: 249 DRIKGPWSAQEDRILTRLVEQYGPRNWSLISRYIKGRSGKSCRLRWCNQLSPTVEHRPFS 428
Query: 62 PEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVDHRPLKRS 121
+ED II AHAR+GN+WATIAR+L GRTDNAVKNHWNSTLKR+ ++ V+
Sbjct: 429 TQEDETIIAAHARYGNRWATIARLLPGRTDNAVKNHWNSTLKRRAKGINVNVNDAD---- 596
Query: 122 ASVGAGCLNPSSP-TGSAMSDPGPPAAISTDAPIAVPAAVVVPATDPATSLSLSLPGFDS 180
N P T ++ PG A T+ AVP P +S
Sbjct: 597 --------NEDDPLTALTLAPPGIANATLTED--AVPDHRASP---------------ES 701
Query: 181 SGSGSGSKQLFGAEFLAVMQEMIRKEVRSYMS 212
+ G F +M+++I +EVR Y+S
Sbjct: 702 AAEG----------FWDMMRDVIAREVREYVS 767
>CO981880
Length = 749
Score = 176 bits (446), Expect = 9e-45
Identities = 81/125 (64%), Positives = 98/125 (77%), Gaps = 2/125 (1%)
Frame = +1
Query: 2 DRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFT 61
+RVKG WSP+ED L +LV HG +NW+ IS IPGRSGKSCRLRWCNQLSPEV+HRPFT
Sbjct: 337 NRVKGSWSPQEDATLLKLVNEHGARNWSVISAGIPGRSGKSCRLRWCNQLSPEVQHRPFT 516
Query: 62 PEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVDHR--PLK 119
P ED II+AHA GNKWATI+R+L GRTDNA+KNHWNSTL+R+ +A ++ P K
Sbjct: 517 PAEDKMIIKAHAIHGNKWATISRLLPGRTDNAIKNHWNSTLRRRRTAXQSSSSSASPPAK 696
Query: 120 RSASV 124
R +S+
Sbjct: 697 RPSSL 711
>BM525863
Length = 439
Score = 126 bits (316), Expect(2) = 5e-37
Identities = 55/72 (76%), Positives = 62/72 (85%)
Frame = +2
Query: 35 IPGRSGKSCRLRWCNQLSPEVEHRPFTPEEDSAIIRAHARFGNKWATIARILNGRTDNAV 94
+PGRSGKSCRLRWCNQL P V+ +PFT EEDS I+ AHA GNKWA IAR+L GRTDNA+
Sbjct: 209 VPGRSGKSCRLRWCNQLDPCVKRKPFTEEEDSIIVSAHAIHGNKWAAIARLLPGRTDNAI 388
Query: 95 KNHWNSTLKRKC 106
KNHWNSTLKR+C
Sbjct: 389 KNHWNSTLKRRC 424
Score = 45.4 bits (106), Expect(2) = 5e-37
Identities = 19/30 (63%), Positives = 23/30 (76%)
Frame = +1
Query: 3 RVKGPWSPEEDEALTRLVQVHGPKNWTAIS 32
RVKGPWSPEED L+RLV G +NW+ I+
Sbjct: 112 RVKGPWSPEEDALLSRLVAQFGARNWSMIA 201
Score = 34.7 bits (78), Expect = 0.040
Identities = 15/45 (33%), Positives = 26/45 (57%)
Frame = +2
Query: 7 PWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQL 51
P++ EED + +HG K W AI++ +PGR+ + + W + L
Sbjct: 281 PFTEEEDSIIVSAHAIHGNK-WAAIARLLPGRTDNAIKNHWNSTL 412
>TC232815 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose-responsive
element binding factor, partial (29%)
Length = 589
Score = 142 bits (358), Expect = 1e-34
Identities = 68/101 (67%), Positives = 77/101 (75%), Gaps = 2/101 (1%)
Frame = +1
Query: 30 AISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPEEDSAIIRAHARFGNKWATIARILNGR 89
+IS+ I GRSGKSCRLRWCNQLSP VEHRPF+ ED I+ AHARFGNKWATIAR+L GR
Sbjct: 10 SISRHIKGRSGKSCRLRWCNQLSPTVEHRPFSTREDEVILHAHARFGNKWATIARMLPGR 189
Query: 90 TDNAVKNHWNSTLKRKCSAVDTAVDHRPLK--RSASVGAGC 128
TDNAVKNHWN+TLKR+ D V+ PL A G+GC
Sbjct: 190 TDNAVKNHWNATLKRRRFGADVVVEDDPLTALTLAPPGSGC 312
>CA784799
Length = 387
Score = 119 bits (299), Expect = 1e-27
Identities = 53/100 (53%), Positives = 67/100 (67%)
Frame = +3
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPEE 64
+G W P EDE L LV+ +GP NW AI++ + GRSGKSCRLRW NQL P + PFT EE
Sbjct: 39 RGHWRPAEDEKLRELVERYGPHNWNAIAEKLRGRSGKSCRLRWFNQLDPRINRSPFTEEE 218
Query: 65 DSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKR 104
+ ++ +H G WA IAR GR+DNAVKNHW+ + R
Sbjct: 219 EERLLASHRIHGFLWAVIARHFPGRSDNAVKNHWHVMMAR 338
>TC229681 UP|Q9S7E3 (Q9S7E3) GmMYB29A1 protein, complete
Length = 795
Score = 117 bits (293), Expect = 5e-27
Identities = 63/138 (45%), Positives = 84/138 (60%), Gaps = 4/138 (2%)
Frame = +1
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG--RSGKSCRLRWCNQLSPEVEHRPFTP 62
KGPW+PEED+ L +Q HG NW A+ K + G R GKSCRLRW N L P+++ F+
Sbjct: 40 KGPWTPEEDQILMSYIQKHGHGNWRALPK-LAGLLRCGKSCRLRWINYLRPDIKRGNFSS 216
Query: 63 EEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDT--AVDHRPLKR 120
EE+ II+ H GN+W+ IA L GRTDN +KN W++ LK++ DT V +KR
Sbjct: 217 EEEEIIIKMHELLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKRLMNSDTNKRVSKPRIKR 396
Query: 121 SASVGAGCLNPSSPTGSA 138
S S + L S PT S+
Sbjct: 397 SDS-NSSTLTQSEPTSSS 447
>TC232084 UP|Q84PP2 (Q84PP2) Transcription factor MYB101 (Fragment), complete
Length = 1383
Score = 115 bits (288), Expect = 2e-26
Identities = 57/115 (49%), Positives = 71/115 (61%), Gaps = 2/115 (1%)
Frame = +2
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIP-GRSGKSCRLRWCNQLSPEVEHRPFTPE 63
KGPW+PEED L +Q HG +W A+ K R GKSCRLRW N L P+++ F+ E
Sbjct: 92 KGPWTPEEDRILVDYIQKHGHGSWRALPKHARLNRCGKSCRLRWTNYLRPDIKRGKFSEE 271
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVD-TAVDHRP 117
E+ II HA GNKW+ IA L GRTDN +KN WN+ LK+K + V HRP
Sbjct: 272 EEQLIINLHAVLGNKWSAIAGHLPGRTDNEIKNFWNTHLKKKLLQMGLDPVTHRP 436
>TC203404 GmMYB16
Length = 1138
Score = 115 bits (288), Expect = 2e-26
Identities = 76/205 (37%), Positives = 101/205 (49%), Gaps = 20/205 (9%)
Frame = +1
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG-RSGKSCRLRWCNQLSPEVEHRPFTPE 63
KGPW+P+ED LT+ +Q HG W ++ K R GKSCRLRW N L P+++ PE
Sbjct: 145 KGPWTPKEDALLTKYIQAHGEGQWKSLPKKAGLLRCGKSCRLRWMNYLRPDIKRGNIAPE 324
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDT------AVDHRP 117
ED IIR H+ GN+W+ IA L GRTDN +KN+WN+ L +K T + P
Sbjct: 325 EDDLIIRMHSLLGNRWSLIAGRLPGRTDNEIKNYWNTHLSKKLKIQGTEDTDTHKMLENP 504
Query: 118 LKRSASVG------------AGCLNPSSPTGSAMSDPGPPAAISTDAPIAVPAAVVVPAT 165
+ +AS G G N G +P P + PI V A+ + T
Sbjct: 505 QEEAASDGGNNNKKKKKKKNGGKKNKQKNKGKENDEP-PKTQVYLPKPIRV-KAMYLQRT 678
Query: 166 DPATSLSLSLPGFDS-SGSGSGSKQ 189
D T FDS S SGS S++
Sbjct: 679 DSNTFT------FDSNSASGSTSQE 735
>TC229682 UP|Q9XIU9 (Q9XIU9) GmMYB29A2 protein, complete
Length = 1249
Score = 115 bits (288), Expect = 2e-26
Identities = 62/138 (44%), Positives = 83/138 (59%), Gaps = 4/138 (2%)
Frame = +3
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG--RSGKSCRLRWCNQLSPEVEHRPFTP 62
KGPW+PEED+ L +Q HG NW A+ K + G R GKSCRLRW N L P+++ F+
Sbjct: 123 KGPWTPEEDQILMSYIQKHGHGNWRALPK-LAGLLRCGKSCRLRWINYLRPDIKRGNFSS 299
Query: 63 EEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKC--SAVDTAVDHRPLKR 120
EE+ II+ H GN+W+ IA L GRTDN +KN W++ LK++ S + V +KR
Sbjct: 300 EEEEIIIKMHELLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKRLLNSDIQKRVSKPRIKR 479
Query: 121 SASVGAGCLNPSSPTGSA 138
S S + L PT SA
Sbjct: 480 SDS-NSSTLTQLEPTSSA 530
>TC217107 homologue to UP|Q7X9I1 (Q7X9I1) MYB transcription factor R2R3 type,
partial (68%)
Length = 1058
Score = 112 bits (281), Expect = 1e-25
Identities = 58/131 (44%), Positives = 79/131 (60%), Gaps = 3/131 (2%)
Frame = +2
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG-RSGKSCRLRWCNQLSPEVEHRPFTPE 63
KG W+ EED+ L ++ HG W ++ K+ R GKSCRLRW N L P+++ FT E
Sbjct: 305 KGAWTKEEDDRLISYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRPDLKRGNFTEE 484
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKC--SAVDTAVDHRPLKRS 121
ED II+ H+ GNKW+ IA L GRTDN +KN+WN+ ++RK +D A HRPL +
Sbjct: 485 EDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKLLNRGIDPAT-HRPLNEA 661
Query: 122 ASVGAGCLNPS 132
A+ N S
Sbjct: 662 ATAATVTTNIS 694
>TC218769 UP|Q9XIU8 (Q9XIU8) GmMYB29A2 protein, complete
Length = 828
Score = 112 bits (281), Expect = 1e-25
Identities = 54/118 (45%), Positives = 72/118 (60%), Gaps = 1/118 (0%)
Frame = +1
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG-RSGKSCRLRWCNQLSPEVEHRPFTPE 63
KGPW+PEED+ LT + HG NW A+ K R GKSCRLRW N L P+++ FT E
Sbjct: 40 KGPWAPEEDQILTSYIDKHGHGNWRALPKQAGLLRCGKSCRLRWINYLRPDIKRGNFTIE 219
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVDHRPLKRS 121
E+ II+ H GN+W+ IA L GRTDN +KN W++ LK++ D + KR+
Sbjct: 220 EEETIIKLHDMLGNRWSAIAAKLPGRTDNEIKNVWHTNLKKRLLKSDQSKSKPSSKRA 393
>TC207828 homologue to UP|Q6SL45 (Q6SL45) MYB6, partial (52%)
Length = 999
Score = 112 bits (279), Expect = 2e-25
Identities = 57/125 (45%), Positives = 77/125 (61%), Gaps = 3/125 (2%)
Frame = +2
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG-RSGKSCRLRWCNQLSPEVEHRPFTPE 63
KG W+ EEDE L +++HG W ++ K+ R GKSCRLRW N L P+++ FT E
Sbjct: 80 KGAWTKEEDERLINYIKLHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRPDLKRGNFTEE 259
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK--CSAVDTAVDHRPLKRS 121
ED II H+ GNKW+ IA L GRTDN +KN+WN+ +KRK +D HRPL +
Sbjct: 260 EDELIINLHSLLGNKWSLIAARLPGRTDNEIKNYWNTHIKRKLYSRGIDPQT-HRPLNAA 436
Query: 122 ASVGA 126
++ A
Sbjct: 437 SATPA 451
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.129 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,162,988
Number of Sequences: 63676
Number of extensions: 154246
Number of successful extensions: 1292
Number of sequences better than 10.0: 218
Number of HSP's better than 10.0 without gapping: 1139
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1197
length of query: 241
length of database: 12,639,632
effective HSP length: 94
effective length of query: 147
effective length of database: 6,654,088
effective search space: 978150936
effective search space used: 978150936
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0321.10


