

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
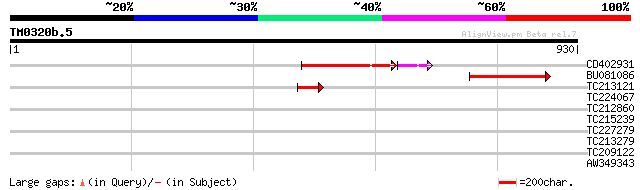
Query= TM0320b.5
(930 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CD402931 similar to GP|9081793|dbj P0489A01.23 {Oryza sativa (ja... 165 3e-46
BU081086 weakly similar to GP|27817864|db OJ1081_B12.13 {Oryza s... 147 3e-35
TC213121 65 1e-10
TC224067 40 0.005
TC212860 weakly similar to UP|PIF1_YEAST (P07271) DNA repair and... 35 0.20
TC215239 UP|H3_PEA (P02300) Histone H3, partial (39%) 33 0.44
TC227279 32 1.3
TC213279 similar to UP|TRSF_DROHY (Q23949) Female-specific trans... 29 8.4
TC209122 similar to UP|Q93VB2 (Q93VB2) AT3g62770/F26K9_200, part... 29 8.4
AW349343 weakly similar to GP|15912305|gb AT4g18900/F13C5_70 {Ar... 29 8.4
>CD402931 similar to GP|9081793|dbj P0489A01.23 {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 617
Score = 165 bits (418), Expect(2) = 3e-46
Identities = 88/156 (56%), Positives = 112/156 (71%)
Frame = +2
Query: 479 GSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGRTAHSRFSIPITIHESSTCNVR 538
G+ KTF+W TLSA +RS+G ++ + +ASLLLP G TAHS FSIP+ I + STCN+
Sbjct: 11 GTSKTFLWKTLSAGMRSKG-LMSSSCLKRLASLLLPRG*TAHSTFSIPLVIKDDSTCNIN 187
Query: 539 QGSHKAEMLQKASLIIWDEAPMLNKHCFEALDKTLNDIMKTQATFGHDKPFAGKVVVLGG 598
QGS ++++L LIIWDE P++NK CFEALD+TL DIM +Q KPF GK VVLGG
Sbjct: 188 QGSARSKLLLHTKLIIWDETPVMNKFCFEALDRTLQDIMASQNKDNATKPFGGK-VVLGG 364
Query: 599 DFRQILPVILKGSRSEIISSSANSSYLWKHCKVMKL 634
DFRQILPVI KGSR +I+ S+ N+S KV+KL
Sbjct: 365 DFRQILPVIRKGSRQDIVGSAINASK-----KVLKL 457
Score = 39.7 bits (91), Expect(2) = 3e-46
Identities = 23/57 (40%), Positives = 32/57 (55%)
Frame = +1
Query: 637 NMRLQQAGSSSSSLEIKEFADWLLQVGDGTIKPIDEDDSIIEIPTYLLVRESDNPLL 693
NMRL ++ +I+EF DW+L + DG +ED+ I+IP LL NPLL
Sbjct: 463 NMRLGTTNNNEDRDDIREFVDWILMIRDGNRD--EEDEGEIDIPKNLL-----NPLL 612
>BU081086 weakly similar to GP|27817864|db OJ1081_B12.13 {Oryza sativa
(japonica cultivar-group)}, partial (7%)
Length = 426
Score = 147 bits (370), Expect = 3e-35
Identities = 74/137 (54%), Positives = 97/137 (70%), Gaps = 3/137 (2%)
Frame = +2
Query: 754 DEDSEIDAEWF---TSEFLNDLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIV 810
++ I++E F T+EF+N L S +PNH+I LKV IML++N+DQ+ GLCN TRLI+
Sbjct: 14 EKSDTIESEGFSTITTEFINSLSTSRLPNHKIKLKVDSRIMLLRNLDQNEGLCNSTRLII 193
Query: 811 SALTPYIIVATALSGSKTGKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQG 870
+ +II A +SG G VYIPRL+ +PS + PFK RR+FPI V +AMTINKSQG
Sbjct: 194 TMFADHIIEAKIMSGKGQGNTVYIPRLATSPSQSPWPFKLIRRKFPIIVSYAMTINKSQG 373
Query: 871 QSLSHVGLYLPRPVFTH 887
Q L+ VGLYLP PVF+H
Sbjct: 374 QLLASVGLYLPTPVFSH 424
>TC213121
Length = 713
Score = 65.5 bits (158), Expect = 1e-10
Identities = 29/44 (65%), Positives = 36/44 (80%)
Frame = +2
Query: 472 FFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPG 515
+FLYG+GG+GKTFVW TLS+ + S IVL +ASSGI S+LLPG
Sbjct: 479 YFLYGYGGTGKTFVWKTLSSTIHSNSGIVLTMASSGILSMLLPG 610
>TC224067
Length = 415
Score = 40.0 bits (92), Expect = 0.005
Identities = 27/48 (56%), Positives = 33/48 (68%), Gaps = 2/48 (4%)
Frame = -2
Query: 859 VCFAMTINKSQGQSLSHV-GLYLPRPV-FTHGQLYVALSRVKSRKRLK 904
VC+ M INKS GQ+LS V G+ LPRP +HGQ YV L++V S R K
Sbjct: 360 VCYTMKINKSHGQTLSQVGGVLLPRPN**SHGQ-YVTLNQV*SY*RRK 220
>TC212860 weakly similar to UP|PIF1_YEAST (P07271) DNA repair and
recombination protein PIF1, mitochondrial precursor,
partial (8%)
Length = 596
Score = 34.7 bits (78), Expect = 0.20
Identities = 21/45 (46%), Positives = 29/45 (63%)
Frame = +2
Query: 861 FAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSRKRLKI 905
+AM+I+K QG +L V L R F G +YVALSRV+S + L +
Sbjct: 5 WAMSIHKCQGMTLERVHTDLSR-AFGCGMVYVALSRVRSLEGLHL 136
>TC215239 UP|H3_PEA (P02300) Histone H3, partial (39%)
Length = 340
Score = 33.5 bits (75), Expect = 0.44
Identities = 13/18 (72%), Positives = 16/18 (88%)
Frame = -3
Query: 855 FPITVCFAMTINKSQGQS 872
FP+ VCFAMT NKS+GQ+
Sbjct: 326 FPLIVCFAMTTNKSEGQT 273
>TC227279
Length = 767
Score = 32.0 bits (71), Expect = 1.3
Identities = 31/79 (39%), Positives = 40/79 (50%), Gaps = 4/79 (5%)
Frame = +2
Query: 458 PRYWMLFR-PITEDFFFLYGFGGS-GKTFVWNTLSAA--LRSEGKIVLNVASSGIASLLL 513
PR ++F+ +ED FL GGS GK VW+TLS A R G N + SG S L
Sbjct: 323 PRAGVIFKISFSEDNPFLLAIGGSKGKLQVWDTLSDAGISRRYGNSNKNRSQSGS*SCSL 502
Query: 514 PGGRTAHSRFSIPITIHES 532
P RTA + +H+S
Sbjct: 503 P-SRTAQ------VVLHKS 538
>TC213279 similar to UP|TRSF_DROHY (Q23949) Female-specific transformer
protein, partial (11%)
Length = 481
Score = 29.3 bits (64), Expect = 8.4
Identities = 23/72 (31%), Positives = 33/72 (44%), Gaps = 1/72 (1%)
Frame = -1
Query: 850 FSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVK-SRKRLKILIV 908
FS + F + +S + L LPR VF H + +AL R SR+ L +LIV
Sbjct: 364 FSLQLFDLLALICTVAPQSVELRRQLLDLLLPRHVFAHQLVALALQRRDLSRRFLAVLIV 185
Query: 909 DDKGVVSNCTRN 920
D V + R+
Sbjct: 184 DAAAVGNTFVRH 149
>TC209122 similar to UP|Q93VB2 (Q93VB2) AT3g62770/F26K9_200, partial (28%)
Length = 692
Score = 29.3 bits (64), Expect = 8.4
Identities = 27/79 (34%), Positives = 36/79 (45%), Gaps = 5/79 (6%)
Frame = -1
Query: 741 ETKYLSYDTPCRSDEDS-EIDAEWFTSEFLNDLKCSGIPNHRIVLKV-GVPIMLIQNIDQ 798
E KYL TP + + D + F SE L DL C P + LK VP+ L +
Sbjct: 266 ELKYLG-STPLKKERDDIAVGLARFASEVLCDLSCPNSPESTLRLKT*TVPLSL-----E 105
Query: 799 SAGLC---NGTRLIVSALT 814
+A C RL +SAL+
Sbjct: 104 TASHCAVGEKARLYISALS 48
>AW349343 weakly similar to GP|15912305|gb AT4g18900/F13C5_70 {Arabidopsis
thaliana}, partial (23%)
Length = 618
Score = 29.3 bits (64), Expect = 8.4
Identities = 29/78 (37%), Positives = 39/78 (49%), Gaps = 3/78 (3%)
Frame = -2
Query: 458 PRYWMLFR-PITEDFFFLYGFGGS-GKTFVWNTLS-AALRSEGKIVLNVASSGIASLLLP 514
PR ++F+ +ED FL GGS GK VW+TLS A + N + SG S LP
Sbjct: 398 PRAGVIFKISFSEDNPFLLAIGGSKGKLQVWDTLSDAGISRRYGNYKNRSQSGS*SCSLP 219
Query: 515 GGRTAHSRFSIPITIHES 532
RTA + +H+S
Sbjct: 218 -SRTAQ------VVLHKS 186
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.344 0.151 0.484
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,610,033
Number of Sequences: 63676
Number of extensions: 535892
Number of successful extensions: 4752
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 3544
Number of HSP's successfully gapped in prelim test: 140
Number of HSP's that attempted gapping in prelim test: 1151
Number of HSP's gapped (non-prelim): 3770
length of query: 930
length of database: 12,639,632
effective HSP length: 106
effective length of query: 824
effective length of database: 5,889,976
effective search space: 4853340224
effective search space used: 4853340224
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0320b.5


