

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.7
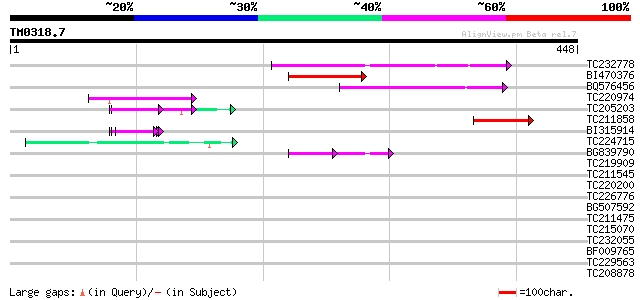
(448 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232778 similar to UP|Q9LNG5 (Q9LNG5) F21D18.16, partial (5%) 73 3e-13
BI470376 weakly similar to GP|14090338|dbj P0638D12.5 {Oryza sat... 55 6e-08
BQ576456 53 3e-07
TC220974 similar to UP|MP62_LYTPI (P91753) Mitotic apparatus pro... 49 5e-06
TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall pr... 44 1e-04
TC211858 44 1e-04
BI315914 similar to PIR|T09854|T098 proline-rich cell wall prote... 43 3e-04
TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%) 42 7e-04
BG839790 similar to PIR|D96521|D96 protein F21D18.16 [imported] ... 36 9e-04
TC219909 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-... 41 0.001
TC211545 weakly similar to UP|Q7XHQ1 (Q7XHQ1) Calcineurin-like p... 41 0.001
TC220200 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific... 40 0.002
TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {A... 37 0.014
BG507592 similar to PIR|S52985|S52 cell wall protein - alfalfa, ... 37 0.014
TC211475 similar to UP|P79122 (P79122) Pinin, partial (5%) 37 0.014
TC215070 similar to UP|Q40336 (Q40336) Proline-rich cell wall pr... 37 0.018
TC232055 similar to UP|O49524 (O49524) Pherophorin - like protei... 37 0.018
BF009765 similar to PIR|E96661|E96 GMP synthase 61700-64653 [im... 37 0.024
TC229563 37 0.024
TC208878 similar to UP|Q39862 (Q39862) Homeobox-leucine zipper p... 37 0.024
>TC232778 similar to UP|Q9LNG5 (Q9LNG5) F21D18.16, partial (5%)
Length = 746
Score = 72.8 bits (177), Expect = 3e-13
Identities = 54/190 (28%), Positives = 83/190 (43%), Gaps = 1/190 (0%)
Frame = -1
Query: 208 YDGDSDSHRRVRELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTIT 267
Y G+ + L+ Q G + + Y +A LI A +ERW ET +FH+ GE TIT
Sbjct: 584 YQGEKQISEEIVPLLRQCGFYWIMKMGYLKINAALISAFIERWRPETHTFHLRCGEATIT 405
Query: 268 LDDVSALLHLPMGSRFYTPGRGERD-ECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGVL 326
L DV LL L P G + + A LC +L+G E E + +
Sbjct: 404 LQDVLILLGLRTDG---APLIGSTNLDWADLCEELLGVRPQEGEIEGSVVKLSWLAHYFS 234
Query: 327 QTRYEAALAEHRYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGA 386
+ E + + R W++ +G +F +KS ++ ++ L D YAWG
Sbjct: 233 HINIDEGNVE-QLQRFIRAWILRFIGGVIFVNKSSS-RVSLRYLQFLRDFE*CSTYAWGP 60
Query: 387 IALATLYDQL 396
LA LY ++
Sbjct: 59 AMLAYLYREM 30
>BI470376 weakly similar to GP|14090338|dbj P0638D12.5 {Oryza sativa
(japonica cultivar-group)}, partial (5%)
Length = 429
Score = 55.1 bits (131), Expect = 6e-08
Identities = 25/62 (40%), Positives = 39/62 (62%)
Frame = -1
Query: 221 LIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMG 280
L++ +GL+ + + + L+ A +ERW ET +FH+ +GE TITL DVS LL +P+
Sbjct: 396 LLQISGLYPIMKLAQLKVNGALVNAFIERWRPETHTFHLKYGEATITLQDVSVLLGIPVD 217
Query: 281 SR 282
R
Sbjct: 216 GR 211
>BQ576456
Length = 424
Score = 52.8 bits (125), Expect = 3e-07
Identities = 41/133 (30%), Positives = 61/133 (45%)
Frame = +1
Query: 261 FGEMTITLDDVSALLHLPMGSRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNRSQT 320
+GE+TITLDD++ LLHLP+ + DE L +L+ S A+
Sbjct: 28 WGELTITLDDMACLLHLPITGALHRFEPLGVDEAVLLLMELLEVSREEARAKIVRAHRAY 207
Query: 321 IRFGVLQTRYEAALAEHRYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVC 380
+R L+ Y++ + A R +L + + TLFA+KS H V + DL
Sbjct: 208 VRLSWLREVYQSRCQARCWIVAVRAYLFHLVDCTLFANKS-ATHVHVVHLKGF*DLC*SG 384
Query: 381 EYAWGAIALATLY 393
Y WG AL +Y
Sbjct: 385 GYGWGVAALVHMY 423
>TC220974 similar to UP|MP62_LYTPI (P91753) Mitotic apparatus protein p62,
partial (7%)
Length = 436
Score = 48.9 bits (115), Expect = 5e-06
Identities = 29/89 (32%), Positives = 39/89 (43%), Gaps = 4/89 (4%)
Frame = +1
Query: 63 PSPTVEIPTVVSPPS----PMVESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGS 118
PSP E PT S S P + + + EE E SSGEE +E EA+S
Sbjct: 85 PSPLDEPPTASSSDSEEEEPQQQQPSSQKNKEEEDEEVSSGEEEEEDEEEEEAASSEEEE 264
Query: 119 DEDSIPPPTVDDDVLPPEQGEQGAQGGEE 147
+++ +PPP V + PP Q E
Sbjct: 265 EDEDLPPPPVSKNPPPPPANPQPQHSSSE 351
>TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall protein,
partial (48%)
Length = 1076
Score = 44.3 bits (103), Expect = 1e-04
Identities = 34/99 (34%), Positives = 38/99 (38%), Gaps = 1/99 (1%)
Frame = -2
Query: 81 ESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPPEQGEQ 140
E GEE GE GEE GE GEE GE G G E S V+ E G +
Sbjct: 532 EGGGEEGGGEAGGGEEGGGEAGGGEEGGGEEGGGEEGGGELSGDGGGVEAGGGD*EGGGE 353
Query: 141 GAQGGEEDLIQRLPPF-PGGPVELSLLTYYADHKAPWAW 178
GG ED + GG E+ PWAW
Sbjct: 352 EGTGGGEDWTGGGDAWGVGGAGEVD---------GPWAW 263
Score = 43.9 bits (102), Expect = 1e-04
Identities = 20/42 (47%), Positives = 22/42 (51%)
Frame = -2
Query: 80 VESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDED 121
V GE+ GEE GEE GE GEE GEA G G E+
Sbjct: 565 VAGGGEKGGGEEGGGEEGGGEAGGGEEGGGEAGGGEEGGGEE 440
Score = 41.6 bits (96), Expect = 7e-04
Identities = 25/73 (34%), Positives = 31/73 (42%), Gaps = 6/73 (8%)
Frame = -2
Query: 81 ESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLP------ 134
+ GEE GEE GE GEE GE GE G G E+ + D +
Sbjct: 547 KGGGEEGGGEEGGGEAGGGEEGGGEAGGGEEGGGEEGGGEEGGGELSGDGGGVEAGGGD* 368
Query: 135 PEQGEQGAQGGEE 147
GE+G GGE+
Sbjct: 367 EGGGEEGTGGGED 329
Score = 40.4 bits (93), Expect = 0.002
Identities = 34/103 (33%), Positives = 41/103 (39%), Gaps = 2/103 (1%)
Frame = +3
Query: 35 ATSQAVEASAPVV--SPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEES 92
A+ V++S P V SPP P PPSP P PPS SS +S S
Sbjct: 309 ASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSSPPPSSPPPSSPPPASPPPS 488
Query: 93 SGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPP 135
S +S SS SS S + + PPP V L P
Sbjct: 489 SPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAVPPPSLTP 617
Score = 38.1 bits (87), Expect = 0.008
Identities = 24/68 (35%), Positives = 27/68 (39%)
Frame = -2
Query: 80 VESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPPEQGE 139
V G GE+ GEE GEE GE GE G G E+ GE
Sbjct: 580 VAGGGVAGGGEKGGGEEGGGEEGGGEAGGGEEGGGEAGGGEEG--------------GGE 443
Query: 140 QGAQGGEE 147
+G GGEE
Sbjct: 442 EG--GGEE 425
Score = 38.1 bits (87), Expect = 0.008
Identities = 31/91 (34%), Positives = 35/91 (38%)
Frame = +3
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSG 104
P SPP P V P P+ P +PP P SS SS SS SS +S
Sbjct: 303 PQASPPPVQSSPPPVPSSPPPSQSPPPASTPP-PSPLSSPPPSSPPPSSPPPSSPPPASP 479
Query: 105 EESSGEASSGMGGSDEDSIPPPTVDDDVLPP 135
SS +S S S PPP PP
Sbjct: 480 PPSSPPPASPPPSSPPPSSPPPFSPPPATPP 572
Score = 36.6 bits (83), Expect = 0.024
Identities = 33/99 (33%), Positives = 39/99 (39%), Gaps = 4/99 (4%)
Frame = +3
Query: 41 EASAPVVS---PPSPMIEVPLVDYPPSPT-VEIPTVVSPPSPMVESSGEESSGEESSGEE 96
+AS P V PP P P PP+ T P PPS SS SS +S
Sbjct: 306 QASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSSPPPSSPPPSSPPPASPPP 485
Query: 97 SSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPP 135
SS +S SS SS S + PPP +PP
Sbjct: 486 SSPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAVPP 602
Score = 36.6 bits (83), Expect = 0.024
Identities = 32/98 (32%), Positives = 36/98 (36%), Gaps = 3/98 (3%)
Frame = +3
Query: 41 EASAPVVSP--PSPMIEVPLVDYPPSPTVEIPTVV-SPPSPMVESSGEESSGEESSGEES 97
+A P SP P+P P V P P P SPP SS SS S
Sbjct: 264 QAQGPSTSPAPPTPQASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSSPPPS 443
Query: 98 SGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPP 135
S SS +S SS S S PPP+ PP
Sbjct: 444 SPPPSSPPPASPPPSSPPPASPPPSSPPPSSPPPFSPP 557
Score = 34.3 bits (77), Expect = 0.12
Identities = 16/37 (43%), Positives = 17/37 (45%)
Frame = -2
Query: 84 GEESSGEESSGEESSGEESSGEESSGEASSGMGGSDE 120
G G GE+ GEE GEE GEA G G E
Sbjct: 583 GVAGGGVAGGGEKGGGEEGGGEEGGGEAGGGEEGGGE 473
Score = 29.6 bits (65), Expect = 2.9
Identities = 26/93 (27%), Positives = 37/93 (38%), Gaps = 9/93 (9%)
Frame = -2
Query: 64 SPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGE---------ESSGEESSGEASSG 114
SP E+ + P + ES+G + + +GEE+ GE E G G A G
Sbjct: 745 SPEGEMVERLGPGAREGESAGANTLEGDGAGEETGGEERGVTVGVNEGGGTAGGGVAGGG 566
Query: 115 MGGSDEDSIPPPTVDDDVLPPEQGEQGAQGGEE 147
+ G E + E+G A GGEE
Sbjct: 565 VAGGGEKG------GGEEGGGEEGGGEAGGGEE 485
Score = 28.5 bits (62), Expect = 6.4
Identities = 22/84 (26%), Positives = 35/84 (41%), Gaps = 14/84 (16%)
Frame = -2
Query: 51 SPMIEVPL---VDYPPSPTVEIPTVV-----------SPPSPMVESSGEESSGEESSGEE 96
SP E+ + +D P ++P + SP MVE G + ES+G
Sbjct: 856 SPKTEIDV*NNMDSKAHPNTKLPIIF*KDHKF*APLSSPEGEMVERLGPGAREGESAGAN 677
Query: 97 SSGEESSGEESSGEASSGMGGSDE 120
+ + +GEE+ GE G +E
Sbjct: 676 TLEGDGAGEETGGEERGVTVGVNE 605
>TC211858
Length = 524
Score = 44.3 bits (103), Expect = 1e-04
Identities = 21/48 (43%), Positives = 30/48 (61%)
Frame = -1
Query: 367 VYWIGMLEDLGRVCEYAWGAIALATLYDQLSQASRRGTAQMGGFTSLL 414
V ++ DLG+ YAWG AL +Y+QL +ASR T Q+ G+ +LL
Sbjct: 464 VVYLDAFRDLGQSGGYAWGVAALVHMYNQLDEASRTTTRQIVGYLTLL 321
>BI315914 similar to PIR|T09854|T098 proline-rich cell wall protein - upland
cotton, partial (37%)
Length = 246
Score = 42.7 bits (99), Expect = 3e-04
Identities = 20/41 (48%), Positives = 20/41 (48%)
Frame = -2
Query: 80 VESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDE 120
VE GEE GE GEE GE GEE GE G G E
Sbjct: 170 VEGGGEEGGGEAGGGEEGGGEAGGGEEGGGEEGGGEEGGGE 48
Score = 42.0 bits (97), Expect = 6e-04
Identities = 19/41 (46%), Positives = 21/41 (50%)
Frame = -2
Query: 81 ESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDED 121
+ GEE G E GEE GE GEE GEA G G E+
Sbjct: 197 KGGGEEGGGVEGGGEEGGGEAGGGEEGGGEAGGGEEGGGEE 75
Score = 41.6 bits (96), Expect = 7e-04
Identities = 19/34 (55%), Positives = 19/34 (55%)
Frame = -2
Query: 84 GEESSGEESSGEESSGEESSGEESSGEASSGMGG 117
GEE GE GEE GEE GEE GE S GG
Sbjct: 128 GEEGGGEAGGGEEGGGEEGGGEEGGGELSGDGGG 27
Score = 40.4 bits (93), Expect = 0.002
Identities = 25/68 (36%), Positives = 28/68 (40%)
Frame = -2
Query: 80 VESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPPEQGE 139
V GE+ GEE G E GEE GE GE G G E+ GE
Sbjct: 215 VAGGGEKGGGEEGGGVEGGGEEGGGEAGGGEEGGGEAGGGEEG--------------GGE 78
Query: 140 QGAQGGEE 147
+G GGEE
Sbjct: 77 EG--GGEE 60
Score = 37.7 bits (86), Expect = 0.011
Identities = 18/41 (43%), Positives = 19/41 (45%)
Frame = -2
Query: 80 VESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDE 120
V G GE+ GEE G E GEE GEA G G E
Sbjct: 230 VAGGGVAGGGEKGGGEEGGGVEGGGEEGGGEAGGGEEGGGE 108
Score = 37.4 bits (85), Expect = 0.014
Identities = 18/39 (46%), Positives = 18/39 (46%)
Frame = -2
Query: 81 ESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSD 119
E GE GEE GEE GEE GE S GG D
Sbjct: 122 EGGGEAGGGEEGGGEEGGGEEGGGELSGDGGGVEAGGGD 6
Score = 31.6 bits (70), Expect = 0.76
Identities = 24/66 (36%), Positives = 28/66 (42%)
Frame = +1
Query: 70 PTVVSPPSPMVESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVD 129
P PPSP+ SS SS SS SS +S SS +S S S PPP+
Sbjct: 16 PASTPPPSPL--SSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSTPPPSSP 189
Query: 130 DDVLPP 135
PP
Sbjct: 190 PPFSPP 207
Score = 31.2 bits (69), Expect = 0.99
Identities = 27/79 (34%), Positives = 31/79 (39%)
Frame = +1
Query: 48 SPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSGEES 107
SPP P PPSP P PPS SS +S SS +S SS S
Sbjct: 7 SPP------PASTPPPSPLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPS 168
Query: 108 SGEASSGMGGSDEDSIPPP 126
+ SS S + PPP
Sbjct: 169 TPPPSSPPPFSPPPATPPP 225
Score = 30.0 bits (66), Expect = 2.2
Identities = 28/87 (32%), Positives = 34/87 (38%)
Frame = +1
Query: 42 ASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEE 101
AS P PPSP+ P PPS PPS +S SS +S SS
Sbjct: 19 ASTP---PPSPLSSPPPSSPPPSSP--------PPSSPPPASPPPSSPPPASPPPSSPPP 165
Query: 102 SSGEESSGEASSGMGGSDEDSIPPPTV 128
S+ SS S + + PPP V
Sbjct: 166 STPPPSSPPPFSPPPATPPPATPPPAV 246
>TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%)
Length = 1270
Score = 41.6 bits (96), Expect = 7e-04
Identities = 43/171 (25%), Positives = 57/171 (33%), Gaps = 3/171 (1%)
Frame = +2
Query: 13 DVGATEDRHRRLHASSRRGDHAATSQAVEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTV 72
+ +E RRL + R ++SQ + +P +P SP P PP +P
Sbjct: 512 ETSPSEGLKRRLTPTR*RRLWLSSSQPSSSCSPAQAPASPTTSSPTTALPP-----LPAS 676
Query: 73 VSPPSPMVESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDV 132
PPSPM S S +S +S S SS S S S P+
Sbjct: 677 SPPPSPMHSPSSSPSPSAPTSPAATSTPPSPSAPSSAATSPSSAASSTSS---PSSSAPS 847
Query: 133 LPPEQGEQGAQGGEEDLIQRLPPF---PGGPVELSLLTYYADHKAPWAWHA 180
PP PP P+EL L T PW W +
Sbjct: 848 SPPSSWPSSP-----------PPLFQHSDSPLELELAT-------PWCWRS 946
>BG839790 similar to PIR|D96521|D96 protein F21D18.16 [imported] -
Arabidopsis thaliana, partial (2%)
Length = 504
Score = 35.8 bits (81), Expect(2) = 9e-04
Identities = 16/39 (41%), Positives = 23/39 (58%)
Frame = -1
Query: 221 LIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHM 259
L+ Q G + + SY +A L+ L+ERW ET +FHM
Sbjct: 261 LLRQCGFY*IMKMSYLKINASLMTVLIERWRPETHTFHM 145
Score = 24.6 bits (52), Expect(2) = 9e-04
Identities = 19/49 (38%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Frame = -3
Query: 256 SFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERD-ECAALCAQLMG 303
S M E TITL DVS LL L + TP G+ + + A C ++ G
Sbjct: 151 SHEMRCEECTITLQDVSXLLGLRVDE---TPLXGQTNLDWAESCERIAG 14
>TC219909 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (21%)
Length = 943
Score = 41.2 bits (95), Expect = 0.001
Identities = 37/111 (33%), Positives = 46/111 (41%), Gaps = 21/111 (18%)
Frame = +1
Query: 37 SQAVEASAPVVSPP----SPMIE----------VPLVDYPP-SPTVEIPTVVS-----PP 76
SQ+ +A+ P VSPP SP + VP V P SP + IPT+ PP
Sbjct: 271 SQSPKANPPTVSPPLPSPSPSVHSKSLSPASSPVPAVGTPAISPAISIPTLAPETGTPPP 450
Query: 77 SPMVESSGEESSGEESSGEESSGEESSGEESSGEAS-SGMGGSDEDSIPPP 126
S SS S G SS S G SS +G S + S + P P
Sbjct: 451 SLGPSSSSPPSPGPSSSSPPSPGPSSSSPPPAGPTSPPSLAPSSNSTAPSP 603
>TC211545 weakly similar to UP|Q7XHQ1 (Q7XHQ1) Calcineurin-like
phosphoesterase-like, partial (11%)
Length = 1127
Score = 40.8 bits (94), Expect = 0.001
Identities = 29/70 (41%), Positives = 38/70 (53%), Gaps = 1/70 (1%)
Frame = +3
Query: 235 YPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSALLHLPMGSRFYTPGRGERD-E 293
Y +A LI L+ER +T +FHM E TITL DVS LL + + TP G + +
Sbjct: 15 YLKINASLITVLIER*RPKTHTFHMRCRECTITLQDVSVLLGMRVDG---TPLIGSTNLD 185
Query: 294 CAALCAQLMG 303
A LC L+G
Sbjct: 186 WADLCV*LLG 215
>TC220200 weakly similar to UP|Q7M4A3 (Q7M4A3) Spermatid-specific protein T2
precursor, partial (40%)
Length = 506
Score = 40.4 bits (93), Expect = 0.002
Identities = 28/94 (29%), Positives = 39/94 (40%), Gaps = 10/94 (10%)
Frame = +2
Query: 34 AATSQAVEASAPVVSP----------PSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESS 83
A + A E+ AP +P PSP ++PPSPT P+ SPP+P S+
Sbjct: 188 ATAATAAESPAPTPTPDFPPSSSPPFPSPASNPSTANFPPSPT-PAPSHHSPPAPSPSSN 364
Query: 84 GEESSGEESSGEESSGEESSGEESSGEASSGMGG 117
S + G +G + S SS GG
Sbjct: 365 PSPSPAPAPVDDAHDGVSHAGVDGSDGKSSSSGG 466
>TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {Arabidopsis
thaliana;} , partial (68%)
Length = 1307
Score = 37.4 bits (85), Expect = 0.014
Identities = 33/101 (32%), Positives = 41/101 (39%), Gaps = 1/101 (0%)
Frame = +2
Query: 36 TSQAVEASAPVVSPPSPMIEVPLVD-YPPSPTVEIPTVVSPPSPMVESSGEESSGEESSG 94
TS A AS + P +P PLV PP+PT P PPS SS +S S+
Sbjct: 557 TSTASSASLRLAFPSAP----PLVSAMPPTPTNAPPPPSPPPSATPPSS---TSPSPSTS 715
Query: 95 EESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPP 135
S+ SS S+ S S P PT + PP
Sbjct: 716 STSAAPSPPSTPSSNTPSARRTSSSTSSSPKPTSNPSSNPP 838
>BG507592 similar to PIR|S52985|S52 cell wall protein - alfalfa, partial
(36%)
Length = 435
Score = 37.4 bits (85), Expect = 0.014
Identities = 19/44 (43%), Positives = 26/44 (58%), Gaps = 6/44 (13%)
Frame = +3
Query: 45 PVVSPP---SPMIEVPLVDYPP---SPTVEIPTVVSPPSPMVES 82
P+V PP P++ P + YPP +P V P VV+PPSP E+
Sbjct: 228 PIVKPPIVYPPVVPTPPIVYPPVVPTPPVVTPPVVTPPSPSSET 359
Score = 30.4 bits (67), Expect = 1.7
Identities = 18/44 (40%), Positives = 24/44 (53%), Gaps = 3/44 (6%)
Frame = +3
Query: 40 VEASAPVVSPP---SPMIEVPLVDYPPSPTVEIPTVVSPPSPMV 80
V + P+V PP +P + P V PPSP+ E P PP P+V
Sbjct: 261 VVPTPPIVYPPVVPTPPVVTPPVVTPPSPSSETP-CPPPPPPVV 389
Score = 28.9 bits (63), Expect = 4.9
Identities = 17/37 (45%), Positives = 20/37 (53%), Gaps = 9/37 (24%)
Frame = +3
Query: 45 PVVSPPSPMIEVP-------LVDYPP--SPTVEIPTV 72
PVV+PPSP E P +V YPP PT I T+
Sbjct: 324 PVVTPPSPSSETPCPPPPPPVVPYPPPAQPTCPIDTL 434
>TC211475 similar to UP|P79122 (P79122) Pinin, partial (5%)
Length = 436
Score = 37.4 bits (85), Expect = 0.014
Identities = 24/74 (32%), Positives = 31/74 (41%), Gaps = 1/74 (1%)
Frame = +2
Query: 63 PSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDED- 121
PSP E PT S S EE ++ EE E SSGE +ED
Sbjct: 53 PSPLDEPPTASS------SDSEEEEEQQQQQPSSQQHEEEEEEVSSGEEEEASSEEEEDE 214
Query: 122 SIPPPTVDDDVLPP 135
++PPP + + PP
Sbjct: 215 NLPPPPISKNPPPP 256
>TC215070 similar to UP|Q40336 (Q40336) Proline-rich cell wall protein,
partial (82%)
Length = 1597
Score = 37.0 bits (84), Expect = 0.018
Identities = 19/44 (43%), Positives = 26/44 (58%), Gaps = 6/44 (13%)
Frame = +2
Query: 45 PVVSPP---SPMIEVPLVDYPP---SPTVEIPTVVSPPSPMVES 82
P+V PP P++ P + YPP +P V P VV+PPSP E+
Sbjct: 740 PIVKPPIVYPPVVPTPPIVYPPVVPTPPVVTPPVVTPPSPPSET 871
Score = 29.6 bits (65), Expect = 2.9
Identities = 18/44 (40%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Frame = +2
Query: 40 VEASAPVVSPP---SPMIEVPLVDYPPSPTVEIPTVVSPPSPMV 80
V + P+V PP +P + P V PPSP E P PP P+V
Sbjct: 773 VVPTPPIVYPPVVPTPPVVTPPVVTPPSPPSETP-CPPPPPPVV 901
Score = 28.5 bits (62), Expect = 6.4
Identities = 17/37 (45%), Positives = 20/37 (53%), Gaps = 9/37 (24%)
Frame = +2
Query: 45 PVVSPPSPMIEVP-------LVDYPP--SPTVEIPTV 72
PVV+PPSP E P +V YPP PT I T+
Sbjct: 836 PVVTPPSPPSETPCPPPPPPVVPYPPPAQPTCPIDTL 946
>TC232055 similar to UP|O49524 (O49524) Pherophorin - like protein, partial
(7%)
Length = 448
Score = 37.0 bits (84), Expect = 0.018
Identities = 27/72 (37%), Positives = 33/72 (45%), Gaps = 3/72 (4%)
Frame = -3
Query: 24 LHASSRRGDHAATSQAVEASAPVVSPPSPMIEVPLVD--YPPSPTVEIPT-VVSPPSPMV 80
L AS RR A + + ASA +PP P + P + PPSP E V PP
Sbjct: 446 LPASLRRSQSLAANPSPSASASGKTPPPPALSAPSPEPLLPPSPEAEAAVEEVPPPHSTQ 267
Query: 81 ESSGEESSGEES 92
SS EE + ES
Sbjct: 266 ASSAEEVAVAES 231
>BF009765 similar to PIR|E96661|E96 GMP synthase 61700-64653 [imported] -
Arabidopsis thaliana, partial (20%)
Length = 412
Score = 36.6 bits (83), Expect = 0.024
Identities = 31/103 (30%), Positives = 38/103 (36%), Gaps = 3/103 (2%)
Frame = +3
Query: 27 SSRRGDHAATSQAVEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGE- 85
SS A+ + AP + PSP P P +PT P PPSP SSG
Sbjct: 60 SSLAASAASRCSPYASPAPPLFQPSPTSTPPSSFSPGAPTPSTPR-TPPPSPTASSSGPI 236
Query: 86 --ESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPP 126
S S+ SS +S S+ E S M PP
Sbjct: 237 PMASPSSASATASSSSFSASAATSASETSRSMAAWKSALTNPP 365
>TC229563
Length = 669
Score = 36.6 bits (83), Expect = 0.024
Identities = 31/119 (26%), Positives = 48/119 (40%), Gaps = 11/119 (9%)
Frame = +1
Query: 48 SPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSGEES 107
+P M LVDY T + +V P P E E+ + EE+ EE E + +E
Sbjct: 7 APAGIMDANKLVDYVYRRTKKQAKIVPQPEPEPEKKEEKPAEEETKPEEEKPPEEAKKEE 186
Query: 108 SGEASSGMGGSDEDS-----------IPPPTVDDDVLPPEQGEQGAQGGEEDLIQRLPP 155
GE GG + +P +DD+ L ++ Q +I+R+PP
Sbjct: 187 GGENKEEKGGEESKEEAKKEENNVVVVPYNNIDDEGL--KRMMYYYQYPPLSVIERIPP 357
>TC208878 similar to UP|Q39862 (Q39862) Homeobox-leucine zipper protein,
partial (46%)
Length = 701
Score = 36.6 bits (83), Expect = 0.024
Identities = 22/68 (32%), Positives = 31/68 (45%)
Frame = +1
Query: 54 IEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSGEESSGEASS 113
I+V L PPS V+ VS P+ + S + S E +G+E+ SS S +
Sbjct: 217 IDVNLPPPPPSAVVDDENAVSSPNSTISSISGKRSEREGNGDENERTSSSRGGGSDDDDG 396
Query: 114 GMGGSDED 121
G G D D
Sbjct: 397 GACGGDGD 420
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.132 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,944,378
Number of Sequences: 63676
Number of extensions: 320530
Number of successful extensions: 3367
Number of sequences better than 10.0: 227
Number of HSP's better than 10.0 without gapping: 2414
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3081
length of query: 448
length of database: 12,639,632
effective HSP length: 100
effective length of query: 348
effective length of database: 6,272,032
effective search space: 2182667136
effective search space used: 2182667136
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0318.7


