

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
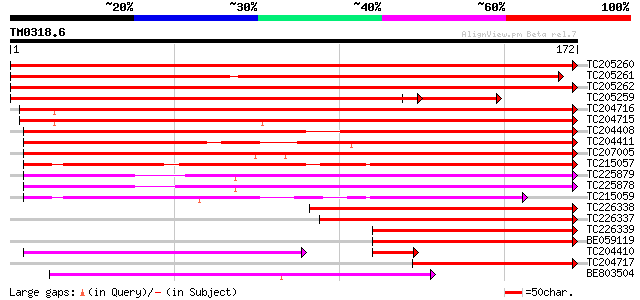
Query= TM0318.6
(172 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205260 similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, part... 314 1e-86
TC205261 similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, part... 297 2e-81
TC205262 weakly similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protei... 263 3e-71
TC205259 similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, part... 218 2e-67
TC204716 similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, part... 233 3e-62
TC204715 similar to UP|Q8H0X0 (Q8H0X0) Expressed protein, partia... 230 3e-61
TC204408 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc f... 164 2e-41
TC204411 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc f... 161 1e-40
TC207005 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc f... 160 2e-40
TC215057 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc f... 145 9e-36
TC225879 similar to GB|AAP88348.1|32815927|BT009714 At3g12630 {A... 133 4e-32
TC225878 similar to GB|AAP88348.1|32815927|BT009714 At3g12630 {A... 130 2e-31
TC215059 similar to UP|Q6T7D0 (Q6T7D0) Fiber protein Fb37, parti... 113 5e-26
TC226338 similar to UP|Q41123 (Q41123) PVPR3 protein, partial (92%) 103 3e-23
TC226337 similar to UP|Q41123 (Q41123) PVPR3 protein, partial (93%) 103 3e-23
TC226339 homologue to UP|Q41123 (Q41123) PVPR3 protein, partial ... 102 1e-22
BE059119 similar to PIR|T11846|T118 pathogenesis-related protein... 88 2e-18
TC204410 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc f... 75 7e-18
TC204717 similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, part... 86 1e-17
BE803504 similar to GP|4510345|gb|A expressed protein {Arabidops... 80 6e-16
>TC205260 similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, partial (71%)
Length = 1037
Score = 314 bits (804), Expect = 1e-86
Identities = 148/172 (86%), Positives = 157/172 (91%)
Frame = +3
Query: 1 MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN 60
ME HDETGCQAPERPILC+NNCGFFGRAATMNMCSKCYKDMLLKQEQDK AA+SVENIVN
Sbjct: 207 MEPHDETGCQAPERPILCINNCGFFGRAATMNMCSKCYKDMLLKQEQDKFAASSVENIVN 386
Query: 61 SCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRV 120
SNGNGKQA+ AV V+VE VEVK V AQ S DSSSG+SLE+KAKTGPSRC TCRKRV
Sbjct: 387 GSSNGNGKQAVATGAVAVQVEAVEVKIVCAQSSVDSSSGDSLEMKAKTGPSRCATCRKRV 566
Query: 121 GLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
GLTGFSCKCGN+FCAMHRYSDKHDCPFDYR VGQ+AIAKANP+IKADKLDKI
Sbjct: 567 GLTGFSCKCGNLFCAMHRYSDKHDCPFDYRTVGQDAIAKANPIIKADKLDKI 722
>TC205261 similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, partial (68%)
Length = 707
Score = 297 bits (760), Expect = 2e-81
Identities = 140/168 (83%), Positives = 152/168 (90%)
Frame = +2
Query: 1 MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN 60
MESHDETGCQAPERPILC+NNCGFFGRAATMNMCSKCYKDMLLKQEQD AA+SVENIVN
Sbjct: 209 MESHDETGCQAPERPILCINNCGFFGRAATMNMCSKCYKDMLLKQEQDNFAASSVENIVN 388
Query: 61 SCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRV 120
SNGNG A+T AV+V+VE VEVK V+AQ S DSSSGESLE+KAK PSRC TCRKRV
Sbjct: 389 GSSNGNG--AVTTGAVDVQVEAVEVKTVSAQSSVDSSSGESLEMKAKNSPSRCATCRKRV 562
Query: 121 GLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADK 168
GLTGFSCKCGN+FCAMHRYSDKH+CPFDYR VGQ+AIAKANP++KADK
Sbjct: 563 GLTGFSCKCGNLFCAMHRYSDKHECPFDYRTVGQDAIAKANPIVKADK 706
>TC205262 weakly similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, partial
(70%)
Length = 838
Score = 263 bits (672), Expect = 3e-71
Identities = 124/172 (72%), Positives = 141/172 (81%)
Frame = +2
Query: 1 MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN 60
ME HDETGCQAPERPILC+NNCGFFGRAATMNMCSK Y DMLLKQE+D+ A++S ENIVN
Sbjct: 323 MEPHDETGCQAPERPILCINNCGFFGRAATMNMCSK*YNDMLLKQERDRFASSSFENIVN 502
Query: 61 SCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRV 120
NGNGK A+ AV V+VE EV V AQ S DS+ G+SLE+KAKTG SRC TCRKRV
Sbjct: 503 GNYNGNGKHAVATGAVAVQVEAAEVNIVCAQSSVDSTYGDSLEMKAKTGTSRCATCRKRV 682
Query: 121 GLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
GLTG SC CGN+FCA+H YSD+HDCP+DYR VGQ+AI ANP+I AD+LD I
Sbjct: 683 GLTGNSC*CGNLFCAVHPYSDEHDCPYDYRTVGQDAITDANPIINADQLDNI 838
>TC205259 similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, partial (44%)
Length = 666
Score = 218 bits (554), Expect(2) = 2e-67
Identities = 105/125 (84%), Positives = 111/125 (88%)
Frame = +3
Query: 1 MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN 60
ME HDETGCQAPERPILC+NNCGFFGRAATMNMCSKCYKDMLLKQEQDK AA+SVENIVN
Sbjct: 219 MEPHDETGCQAPERPILCINNCGFFGRAATMNMCSKCYKDMLLKQEQDKFAASSVENIVN 398
Query: 61 SCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRV 120
SNGNGKQA+ AV V+VE VEVK V AQ S DSSSG+SLE+KAKTGPSRC TCRKRV
Sbjct: 399 GSSNGNGKQAVATGAVAVQVEAVEVKIVCAQSSVDSSSGDSLEMKAKTGPSRCATCRKRV 578
Query: 121 GLTGF 125
GLTGF
Sbjct: 579 GLTGF 593
Score = 55.1 bits (131), Expect(2) = 2e-67
Identities = 21/30 (70%), Positives = 25/30 (83%)
Frame = +1
Query: 120 VGLTGFSCKCGNVFCAMHRYSDKHDCPFDY 149
+G G CKCGN+FCAMHRYSDKH+ PF+Y
Sbjct: 577 LG*LGSXCKCGNLFCAMHRYSDKHEWPFEY 666
>TC204716 similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, partial (67%)
Length = 1331
Score = 233 bits (595), Expect = 3e-62
Identities = 107/170 (62%), Positives = 133/170 (77%), Gaps = 1/170 (0%)
Frame = +1
Query: 4 HDETGCQAP-ERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSC 62
HD+TGCQAP E PILC+NNCGFFG AAT++MCSKC+KDM+LKQEQ KL A+S+ NI+N
Sbjct: 346 HDKTGCQAPPEGPILCINNCGFFGSAATLSMCSKCHKDMMLKQEQAKLVASSIGNIMNGS 525
Query: 63 SNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGL 122
S+ +G + + A +V+V V VE K ++AQ S S ES E K K GP RC C KRVGL
Sbjct: 526 SSSSGNEPVVATSVDVSVNSVESKIISAQPLVASGSDESDEAKPKDGPKRCSNCNKRVGL 705
Query: 123 TGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
TGF+C+CGN+FC+ HRYSDKH+CPFDYR ++AIAKANP +KA+KLDKI
Sbjct: 706 TGFNCRCGNLFCSEHRYSDKHNCPFDYRTAARDAIAKANPTVKAEKLDKI 855
>TC204715 similar to UP|Q8H0X0 (Q8H0X0) Expressed protein, partial (76%)
Length = 1150
Score = 230 bits (586), Expect = 3e-61
Identities = 111/173 (64%), Positives = 134/173 (77%), Gaps = 4/173 (2%)
Frame = +2
Query: 4 HDETGCQAP-ERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSC 62
HD+TGCQAP E PILC+NNCGFFG AATMNMCSKC+KDMLLKQEQ KLAA+S+ NI+N
Sbjct: 188 HDKTGCQAPPEGPILCINNCGFFGSAATMNMCSKCHKDMLLKQEQAKLAASSIGNIMNGS 367
Query: 63 SNGNGKQAITADA---VNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKR 119
S+ K+ + A A V++ V PVE K V+ Q S S E K K GP RC +C KR
Sbjct: 368 SSSTEKEPVVAAAAANVDIPVIPVEPKTVSVQPLFGSGPEGSGEAKLKDGPKRCSSCNKR 547
Query: 120 VGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
VGLTGF+C+CG++FCA+HRYSDKH+CPFDYR ++AIAKANPV+KA+KLDKI
Sbjct: 548 VGLTGFNCRCGDLFCAVHRYSDKHNCPFDYRTAARDAIAKANPVVKAEKLDKI 706
>TC204408 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc finger-like
protein), partial (68%)
Length = 1097
Score = 164 bits (414), Expect = 2e-41
Identities = 79/168 (47%), Positives = 106/168 (63%)
Frame = +3
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E CQAPE LC NNCGFFG ATMN+CSKCY+D+ LK+E+ +++E ++ S+
Sbjct: 282 EEHRCQAPEGHRLCSNNCGFFGSPATMNLCSKCYRDIRLKEEEQAKTKSTIETALSGSSS 461
Query: 65 GNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTG 124
A D++ VE + +V + S +V A +RCG CRKRVGLTG
Sbjct: 462 ATVAVASAVDSLPAPVESLPQPSVVS----------SPDVAAPVQANRCGACRKRVGLTG 611
Query: 125 FSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
F C+CG FC HRY +KH C FD++AVG+E IA+ANPVIK +KL +I
Sbjct: 612 FKCRCGTTFCGTHRYPEKHACGFDFKAVGREEIARANPVIKGEKLRRI 755
>TC204411 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc finger-like
protein), partial (68%)
Length = 1346
Score = 161 bits (408), Expect = 1e-40
Identities = 81/177 (45%), Positives = 109/177 (60%), Gaps = 9/177 (5%)
Frame = +2
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E CQAPE LC NNCGFFG ATMN+CSKCY+D+ LK+E+ +++E ++
Sbjct: 377 EEHRCQAPEGHRLCSNNCGFFGSPATMNLCSKCYRDIRLKEEEQAKTKSTIETALS---- 544
Query: 65 GNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESL---------EVKAKTGPSRCGT 115
G+ +TA AV V + + + S+ ESL ++ A +RCG
Sbjct: 545 GSSSATVTATAV-----------VASSVESPSAPVESLPQPPVLISPDIAAPVQANRCGA 691
Query: 116 CRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
CRKRVGLTGF C+CG FC HRY +KH C FD++AVG+E IA+ANPVIK +KL +I
Sbjct: 692 CRKRVGLTGFKCRCGTTFCGSHRYPEKHACGFDFKAVGREEIARANPVIKGEKLRRI 862
>TC207005 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc finger-like
protein), partial (73%)
Length = 1085
Score = 160 bits (406), Expect = 2e-40
Identities = 79/175 (45%), Positives = 111/175 (63%), Gaps = 7/175 (4%)
Frame = +2
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E C+ PE LCVNNCGFFG ATMN+CSKCY + LK++++ +++E ++S S+
Sbjct: 212 EEHRCEPPEGHRLCVNNCGFFGSTATMNLCSKCYSAIRLKEQEEASTKSTIETALSSASS 391
Query: 65 GNGKQAITA--DAVNVRVEP-----VEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCR 117
+ + AV+V +E EV+ A +SS S+ + P+RC TCR
Sbjct: 392 AKPSSSTSPPPSAVDVLMESPPPSAAEVEVAVTVTVAVASSSISINSGSVAQPNRCATCR 571
Query: 118 KRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
KRVGLTGF C+CG FC HRY ++H C FD++ VG+E IA+ANPVIKA+KL +I
Sbjct: 572 KRVGLTGFKCRCGVTFCGAHRYPEEHACGFDFKTVGREEIARANPVIKAEKLRRI 736
>TC215057 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc finger-like
protein), partial (63%)
Length = 973
Score = 145 bits (366), Expect = 9e-36
Identities = 74/168 (44%), Positives = 103/168 (61%)
Frame = +1
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E CQAP C NNCGFFG AT NMCSKCY+D LK++Q +++ + ++N
Sbjct: 244 EEHRCQAPR---FCANNCGFFGSPATQNMCSKCYRDFQLKEQQ----SSNAKMVLNQSLV 402
Query: 65 GNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTG 124
+ A+ + + V+ + D + ES EVKA +RC TCR+RVGLTG
Sbjct: 403 PSPPPAVISQPSSSSSAAVDPSSAVV----DDAPRESEEVKAPQ-QNRCMTCRRRVGLTG 567
Query: 125 FSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
F C+CG + C HRY ++H C FD++ +G+E IAKANPV+K +KLDKI
Sbjct: 568 FKCRCGMMLCGTHRYPEQHACEFDFKGMGREQIAKANPVVKGEKLDKI 711
>TC225879 similar to GB|AAP88348.1|32815927|BT009714 At3g12630 {Arabidopsis
thaliana;} , partial (57%)
Length = 1018
Score = 133 bits (335), Expect = 4e-32
Identities = 68/169 (40%), Positives = 94/169 (55%), Gaps = 1/169 (0%)
Frame = +3
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+ET + PE LCVNNCG G AT NMC KC+ + S +
Sbjct: 468 EETDFKVPETITLCVNNCGVTGNPATNNMCQKCF---------------TASTATTSGAG 602
Query: 65 GNG-KQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLT 123
G G T V+ R + + ++ SS ++ +AK +RC CR++VGLT
Sbjct: 603 GAGIASPATRSGVSARPQKRSFPEEPSPVADPPSSDQTTPSEAKRVVNRCSGCRRKVGLT 782
Query: 124 GFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
GF C+CG +FCA HRYSD+HDC +DY+A G+EAIA+ NPVI+A K+ K+
Sbjct: 783 GFRCRCGELFCAEHRYSDRHDCSYDYKAAGREAIARENPVIRAAKIVKV 929
>TC225878 similar to GB|AAP88348.1|32815927|BT009714 At3g12630 {Arabidopsis
thaliana;} , partial (66%)
Length = 995
Score = 130 bits (328), Expect = 2e-31
Identities = 68/169 (40%), Positives = 93/169 (54%), Gaps = 1/169 (0%)
Frame = +3
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+ET + PE LCVNNCG G AT NMC KC+ A + S +
Sbjct: 177 EETDFKVPETITLCVNNCGVTGNPATNNMCQKCF------------TAFTTSTATTSGAG 320
Query: 65 GNG-KQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLT 123
G G T ++ R + + SS ++ +AK +RC CR++VGLT
Sbjct: 321 GAGIASPATRSGISARPLKRSFPEEPSPPADPPSSDQTTPSEAKRVVNRCSGCRRKVGLT 500
Query: 124 GFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
GF C+CG +FCA HRYSD+HDC +DY+A G+EAIA+ NPVI+A K+ K+
Sbjct: 501 GFRCRCGELFCAEHRYSDRHDCSYDYKAAGREAIARENPVIRAAKIVKV 647
>TC215059 similar to UP|Q6T7D0 (Q6T7D0) Fiber protein Fb37, partial (63%)
Length = 710
Score = 113 bits (282), Expect = 5e-26
Identities = 63/162 (38%), Positives = 89/162 (54%), Gaps = 9/162 (5%)
Frame = +2
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVE-------- 56
+E CQAP C NNCGFFG AT NMCSKCY+D LK++Q A +
Sbjct: 287 EEHRCQAPR---FCANNCGFFGSPATHNMCSKCYRDFQLKEQQSSNAKMVLNQSLVPPPP 457
Query: 57 -NIVNSCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGT 115
++++ S+ + A +A AV V A +A+ ++KA +RC T
Sbjct: 458 PSVISQPSSSSAPAADSASAV----------VVDAPRAAE-------DLKAPQ-QNRCMT 583
Query: 116 CRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAI 157
CR+RVGLTGF C+CG + C HRY ++H C FD++ +G+E I
Sbjct: 584 CRRRVGLTGFKCRCGMMLCGTHRYPEQHACEFDFKGMGREQI 709
>TC226338 similar to UP|Q41123 (Q41123) PVPR3 protein, partial (92%)
Length = 860
Score = 103 bits (258), Expect = 3e-23
Identities = 42/81 (51%), Positives = 61/81 (74%)
Frame = +1
Query: 92 ISADSSSGESLEVKAKTGPSRCGTCRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRA 151
+ +S + ++ + K +RC CR+RVGLTGF C+CG++FCA HRYSD+HDC +DY+A
Sbjct: 295 LDEESQTDQTTSSEPKRAVNRCSGCRRRVGLTGFRCRCGDLFCAEHRYSDRHDCSYDYKA 474
Query: 152 VGQEAIAKANPVIKADKLDKI 172
G+EAIA+ NPV+KA K+ K+
Sbjct: 475 AGREAIARENPVVKAAKIVKV 537
>TC226337 similar to UP|Q41123 (Q41123) PVPR3 protein, partial (93%)
Length = 852
Score = 103 bits (258), Expect = 3e-23
Identities = 43/78 (55%), Positives = 60/78 (76%)
Frame = +2
Query: 95 DSSSGESLEVKAKTGPSRCGTCRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQ 154
+S + ++ + K SRC CR+RVGLTGF C+CG++FCA HRYSD+HDC +DY+A G+
Sbjct: 377 ESQTDQTASSEPKRVVSRCSCCRRRVGLTGFRCRCGDLFCAEHRYSDRHDCSYDYKAAGR 556
Query: 155 EAIAKANPVIKADKLDKI 172
EAIA+ NPV+KA K+ K+
Sbjct: 557 EAIARENPVVKAAKIVKV 610
>TC226339 homologue to UP|Q41123 (Q41123) PVPR3 protein, partial (82%)
Length = 750
Score = 102 bits (253), Expect = 1e-22
Identities = 41/62 (66%), Positives = 53/62 (85%)
Frame = +1
Query: 111 SRCGTCRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLD 170
SRC CR+RVGLTGF C+CG++FCA HRYSD+HDC +DY+A G+EAIA+ NPV+KA K+
Sbjct: 286 SRCSCCRRRVGLTGFRCRCGDLFCAEHRYSDRHDCSYDYKAAGREAIARENPVVKAAKIV 465
Query: 171 KI 172
K+
Sbjct: 466 KV 471
>BE059119 similar to PIR|T11846|T118 pathogenesis-related protein 3 - kidney
bean, partial (50%)
Length = 378
Score = 88.2 bits (217), Expect = 2e-18
Identities = 34/62 (54%), Positives = 50/62 (79%)
Frame = +3
Query: 111 SRCGTCRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLD 170
+RC CR++VGLTGF C+CG +FCA HRYSD+H+C ++Y+A G++ IA+ NP I+A K+
Sbjct: 174 NRCSGCRRKVGLTGFRCQCGKLFCAKHRYSDRHNCSYNYKAAGKKPIARENPGIRAAKIV 353
Query: 171 KI 172
K+
Sbjct: 354 KV 359
>TC204410 similar to UP|Q9SJM6 (Q9SJM6) Expressed protein (Zinc finger-like
protein), partial (45%)
Length = 471
Score = 75.1 bits (183), Expect(2) = 7e-18
Identities = 35/86 (40%), Positives = 52/86 (59%)
Frame = +3
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E CQAPE LC NNCGFFG ATMN+CSKCY+D+ LK+E+ +++E ++ S+
Sbjct: 138 EEHRCQAPEGHRLCSNNCGFFGSPATMNLCSKCYRDIRLKEEEQAKTKSTIETALSGSSS 317
Query: 65 GNGKQAITADAVNVRVEPVEVKAVTA 90
A D++ VE + +V +
Sbjct: 318 ATVAVASAVDSLPAPVESLPQPSVVS 395
Score = 31.6 bits (70), Expect(2) = 7e-18
Identities = 12/14 (85%), Positives = 13/14 (92%)
Frame = +2
Query: 111 SRCGTCRKRVGLTG 124
+RCG CRKRVGLTG
Sbjct: 428 NRCGACRKRVGLTG 469
>TC204717 similar to UP|Q6VAG2 (Q6VAG2) Zinc-finger protein, partial (29%)
Length = 544
Score = 85.5 bits (210), Expect = 1e-17
Identities = 33/50 (66%), Positives = 43/50 (86%)
Frame = +1
Query: 123 TGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
T F+C+CGN+FC++HRYSDKH+CPFDY ++AIAKANP +K +KLDKI
Sbjct: 10 TRFNCRCGNLFCSVHRYSDKHNCPFDYHTAARDAIAKANPAVKVEKLDKI 159
>BE803504 similar to GP|4510345|gb|A expressed protein {Arabidopsis
thaliana}, partial (14%)
Length = 394
Score = 79.7 bits (195), Expect = 6e-16
Identities = 40/119 (33%), Positives = 61/119 (50%), Gaps = 2/119 (1%)
Frame = +2
Query: 13 ERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSNGNGKQAIT 72
E ILC NNCGFF MN+CSKCY +LL ++++ L +++E ++S S
Sbjct: 8 ECDILCANNCGFFCSTTPMNLCSKCYGSILLNEQEESLTKSTIETALSSSSMNPTFYTSP 187
Query: 73 ADAVNVRVE--PVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTGFSCKC 129
V+V +E + V ++ D++S + P+ C TCRK LT F C+C
Sbjct: 188 PTLVDVLIESPSPSLAEVAVTVAVDATSNVCTTYRLVAQPNLCATCRKLESLTWFKCRC 364
Score = 28.1 bits (61), Expect = 2.1
Identities = 11/27 (40%), Positives = 18/27 (65%)
Frame = -1
Query: 119 RVGLTGFSCKCGNVFCAMHRYSDKHDC 145
+VGL+ S C N+ +HR+SD++ C
Sbjct: 313 KVGLSNQSVGCANITGRVHRHSDRNLC 233
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.131 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,443,668
Number of Sequences: 63676
Number of extensions: 117905
Number of successful extensions: 625
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 607
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 611
length of query: 172
length of database: 12,639,632
effective HSP length: 91
effective length of query: 81
effective length of database: 6,845,116
effective search space: 554454396
effective search space used: 554454396
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0318.6


