

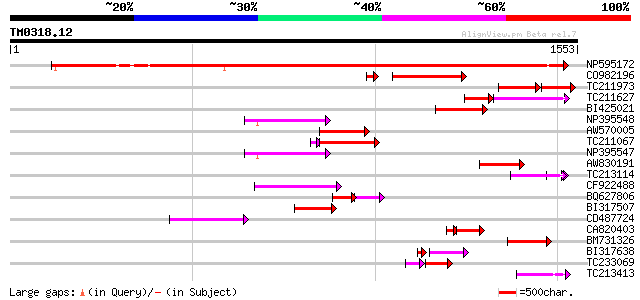
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.12
(1553 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 1174 0.0
CO982196 243 3e-71
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 150 7e-59
TC211627 129 3e-50
BI425021 166 1e-40
NP395548 reverse transcriptase [Glycine max] 151 2e-36
AW570005 151 2e-36
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 145 3e-36
NP395547 reverse transcriptase [Glycine max] 147 5e-35
AW830191 144 4e-34
TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, parti... 127 4e-29
CF922488 125 2e-28
BQ627806 73 8e-28
BI317507 122 1e-27
CD487724 121 2e-27
CA820403 weakly similar to GP|13273463|gb| pol protein integrase... 96 2e-25
BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza... 107 4e-23
BI317638 weakly similar to GP|9294238|dbj| contains similarity t... 92 1e-21
TC233069 84 6e-21
TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 99 1e-20
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 1174 bits (3036), Expect = 0.0
Identities = 633/1440 (43%), Positives = 895/1440 (61%), Gaps = 22/1440 (1%)
Frame = +1
Query: 114 ANAQTERWR--------KLDIPVFQGEEDAYSWIQKLERYFRLKGATEEEKVQAIMVALD 165
+N+Q E+ R KLD P F G+ + WI K E++F + +++ V LD
Sbjct: 244 SNSQKEQQRSSFQVRSVKLDFPRFDGK-NVMDWIFKAEQFFDYYATPDADRLIIASVHLD 420
Query: 166 GKALSWYQWWETCNQVTTWTKFKEAMLERFQLTSNSSPFAALLALKQEGSVEEFVGQFER 225
+ WYQ + ++W F A+ F ++ P A L L Q +V E+ QF
Sbjct: 421 QDVVPWYQMLQKTEPFSSWQAFTRALELDFGPSAYDCPRATLFKLNQSATVNEYYMQFTA 600
Query: 226 FAGMLKGIDEEHYMDIFVNGLKEEIAAEIKLYEPKSLTIMVKKALMVEQKNLAVSKAGIG 285
+ G+ E +D FV+GL+EEI+ ++K EP++LT V A + E+K + K
Sbjct: 601 LVNRVDGLSAEAILDCFVSGLQEEISRDVKAMEPRTLTKAVALAKLFEEKYTSPPKTKTF 780
Query: 286 STSRYNSSFKPPFRSTTFQKSVQIDTGQKSGSGSVTQGTVQSSSGSVNERVGGKP--ARG 343
S N F S T T QK+ + + + + KP R
Sbjct: 781 SNLARN------FTSNTSATQKYPPTNQKNDNPKPNLPPLLPTPST-------KPFNLRN 921
Query: 344 GVYRKLTGEEMKEKLRKGECFRCDEKFGPNHICKNKQFRVMLLR-DDGEEEEIETLEPLT 402
+K++ E++ + K C+ CDEKF P H C N+Q VMLL+ ++ +E++ + +T
Sbjct: 922 QNIKKISPAEIQLRREKNLCYFCDEKFSPAHKCPNRQ--VMLLQLEETDEDQTDEQVMVT 1095
Query: 403 EEIEEAMELKHLSLNSIQGISSRKSLKVWGKLKGTAIVVLVDCGATHNFISQSLAREMEL 462
EE + HLSLN+++G + +++ G++ G A+ +LVD G++ NFI +A+ ++L
Sbjct: 1096 EEANMDDDTHHLSLNAMRGSNGVGTIRFTGQVGGIAVKILVDGGSSDNFIQPRVAQVLKL 1275
Query: 463 QITETPAYTVEVGDGHVVKSKGVCKNLSVQIQGFVVTQDFFLFGLRGVDVVLGLEWLAGL 522
+ P V VG+G ++ ++G+ + L + IQG V +L + G DV+LG WLA L
Sbjct: 1276 PVEPAPNLRVLVGNGQILSAEGIVQQLPLHIQGQEVKVPVYLLQISGADVILGSTWLATL 1455
Query: 523 GDIKANFEELTLKFKLGNKKVVLKGEPELLKKRASMNALVKAIQQQGEGYVLQYQMLEKE 582
G A++ LTLKF +K + L+GE +A ++ + + Q+++KE
Sbjct: 1456 GPHVADYAALTLKFFQNDKFITLQGEGNSEATQAQLHHFRRLQNTKSIEECFAIQLIQKE 1635
Query: 583 VAK-------AEETPELLKAVLSDFPALFAAPTELPPQRRHDHAIHLKEGASIPNIRPYR 635
V + PEL +L + +FA P LPPQR DHAI LK+G+ +RPYR
Sbjct: 1636 VPEDTLKDLPTNIDPELA-ILLHTYAQVFAVPASLPPQREQDHAIPLKQGSGPVKVRPYR 1812
Query: 636 YPHYQKNEIEKLVAEMLNAGIIRPSISPYSSPVILVRKKDGSWRFCVDYRALNKITVPNK 695
YPH QK++IEK++ EML GII+PS SP+S P++LV+KKDGSWRFC DYRALN ITV +
Sbjct: 1813 YPHTQKDQIEKMIQEMLVQGIIQPSNSPFSLPILLVKKKDGSWRFCTDYRALNAITVKDS 1992
Query: 696 FPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLVMPFGLT 755
FP+P +DELLDE+ A+ FSK+DL+SGYHQI+++ ED EKTAFRTH GHYE+LVMPFGLT
Sbjct: 1993 FPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLT 2172
Query: 756 NAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQHDLKINGK 815
NAP+TFQ LMN++ + LRK LVFFDDILIYS + ++H+ HL VLQ + QH L
Sbjct: 2173 NAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLS 2352
Query: 816 KSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWPVPKDLKSLRGFLGLTGYYRRFVKDY 875
K FG +++YLGH +S V+ + K++A+ WP P ++K LRGFLGLTGYYRRF+K Y
Sbjct: 2353 KCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFLGLTGYYRRFIKSY 2532
Query: 876 GKIARPLTQLLKKDSFQWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETDASGTGLG 935
IA PLT LL+KDSF W + AF LK +TE P L++PDFS+ F+LETDASG G+G
Sbjct: 2533 ANIAGPLTDLLQKDSFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQPFILETDASGIGVG 2712
Query: 936 AVLTQEGKPLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLGRHFIIRTDQKSLK 995
AVL Q G P+A++S L+ R Q +S Y REL+A+ A+ ++RHYLLG FIIRTDQ+SLK
Sbjct: 2713 AVLGQNGHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNKFIIRTDQRSLK 2892
Query: 996 FLTEQQVVGEGQFKWISKLSGYDFEIQYKPGKDNSAADAMSRRSSYCALSVLKVNDFEEW 1055
L +Q + Q W+ K GYDF+I+YKPGKDN AADA+S R A S EE
Sbjct: 2893 SLMDQSLQTPEQQAWLHKFLGYDFKIEYKPGKDNQAADALS-RMFMLAWSEPHSIFLEEL 3069
Query: 1056 QTEMLQDPKLQAIVHDLIVDPEAHKGYTLKDHRLFYKGKLVLSKQSSRIPILCKEFHASL 1115
+ ++ DP L+ ++ +A YT+++ L++K ++V+ + + + +E+H+S
Sbjct: 3070 RARLISDPHLKQLMETYKQGADA-SHYTVREGLLYWKDRVVIPAEEEIVNKILQEYHSSP 3246
Query: 1116 LGGHSGFFRTYRRLAAVVYWEGMKSDIRDFVAECDTCQRNKFDNLSPAGLLQPLPVPTQV 1175
+GGH+G RT RL A YW M+ D++ ++ +C CQ+ K +N PAGLLQPLP+P QV
Sbjct: 3247 IGGHAGITRTLARLKAQFYWPKMQEDVKAYIQKCLICQQAKSNNTLPAGLLQPLPIPQQV 3426
Query: 1176 WTDVSMDFIGGLPKSNGKDTILVVVDRLTKYAHFIPLRHPFTAPEVAALFLHEVVRLHGF 1235
W DV+MDFI GLP S G I+VV+DRLTKYAHFIPL+ + + VA F+ +V+LHG
Sbjct: 3427 WEDVAMDFITGLPNSFGLSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSHIVKLHGI 3606
Query: 1236 PTTIVSDRDSLFLSSFWKELFRVSGTQLKFSSAYHPQTDGQTEVVNRCLEVYLRCLTGSR 1295
P +IVSDRD +F S+FW+ LF++ GT L SSAYHPQ+DGQ+EV+N+CLE+YLRC T
Sbjct: 3607 PRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYLRCFTYEH 3786
Query: 1296 PRKWVTCLPWAEFWFNSNYNRSAKMSPFQALYGREPPVLLQGTTIPSKIAAVNDLQVGRD 1355
P+ WV LPWAEFW+N+ Y+ S M+PF+ALYGREPP L + A V + RD
Sbjct: 3787 PKGWVKALPWAEFWYNTAYHMSLGMTPFRALYGREPPTLTRQACSIDDPAEVREQLTDRD 3966
Query: 1356 ELLSDLRANLLKSQDMMRAYANKKRRDVDYQIGDEVFLKLQPYRRRSLAKKMNEKLSPRY 1415
LL+ L+ NL ++Q +M+ A+KKR DV +QIGDEV +KLQPYR+ S + N+KLS RY
Sbjct: 3967 ALLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQPYRQHSAVLRKNQKLSMRY 4146
Query: 1416 YGPYPIVAKIGAVAYRLELPAHSRVHPVFHVSLLKTAVGTNFQPQ-PLPAALNEDHELLV 1474
+GP+ ++AKIG VAY+LELP+ +R+HPVFHVS LK GT P PLP + E ++
Sbjct: 4147 FGPFKVLAKIGDVAYKLELPSAARIHPVFHVSQLKPFNGTAQDPYLPLPLTVTEMGPVM- 4323
Query: 1475 EPESVLAVRETTMG--QIE-VLIQWHSLPACENSWESAAQLHEAFPDFPLEDKVKHLGGG 1531
+P +LA R G QIE +L+QW + E +WE + ++P F LEDKV G G
Sbjct: 4324 QPVKILASRIIIRGHNQIEQILVQWENGLQDEATWEDIEDIKASYPTFNLEDKVVFKGEG 4503
>CO982196
Length = 812
Score = 243 bits (620), Expect(2) = 3e-71
Identities = 116/204 (56%), Positives = 149/204 (72%)
Frame = +1
Query: 1048 KVNDFEEWQTEMLQDPKLQAIVHDLIVDPEAHKGYTLKDHRLFYKGKLVLSKQSSRIPIL 1107
K + +W+ E+ +L I ++ GY ++ +L++K +LVLSK S++IP+L
Sbjct: 199 KRRELADWEEEIQAYLELYEIYQGILTKTTKKPGYAIRGGKLYFKDRLVLSKNSTKIPLL 378
Query: 1108 CKEFHASLLGGHSGFFRTYRRLAAVVYWEGMKSDIRDFVAECDTCQRNKFDNLSPAGLLQ 1167
KE S LGGHSGFFRT++R+A VV+W+GMK RD+VA C+ C+RNK LSPAGLL
Sbjct: 379 LKELQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYVAACEICRRNKTSTLSPAGLL* 558
Query: 1168 PLPVPTQVWTDVSMDFIGGLPKSNGKDTILVVVDRLTKYAHFIPLRHPFTAPEVAALFLH 1227
LP+PT+VWTD+SMDFIGGLPK+ GKD ILVVVDRLTKYAHF L HP+TA EVA LF+
Sbjct: 559 LLPIPTKVWTDISMDFIGGLPKAQGKDNILVVVDRLTKYAHFFALSHPYTAKEVAELFIK 738
Query: 1228 EVVRLHGFPTTIVSDRDSLFLSSF 1251
E+VRLHGFP +IVSD LF+S F
Sbjct: 739 ELVRLHGFPASIVSDXXRLFMSLF 810
Score = 46.2 bits (108), Expect(2) = 3e-71
Identities = 20/34 (58%), Positives = 27/34 (78%)
Frame = +2
Query: 977 RHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQFKW 1010
RHY +G+ FIIRT+ +S KFL EQ+++ E QFKW
Sbjct: 53 RHYPVGKKFIIRTN*RSSKFLNEQRLMSEEQFKW 154
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 150 bits (380), Expect(2) = 7e-59
Identities = 70/113 (61%), Positives = 92/113 (80%)
Frame = +1
Query: 1340 IPSKIAAVNDLQVGRDELLSDLRANLLKSQDMMRAYANKKRRDVDYQIGDEVFLKLQPYR 1399
IPS++ VN L + RD LL+ LR NLLKS D+M A NK+RRD++Y +GD VFLK+QPYR
Sbjct: 1 IPSRLEEVNKLIIARDGLLATLRENLLKS*DIM*ANTNKRRRDIEYVVGD*VFLKMQPYR 180
Query: 1400 RRSLAKKMNEKLSPRYYGPYPIVAKIGAVAYRLELPAHSRVHPVFHVSLLKTA 1452
RRSLAK++NEKLSPR+Y P+ + K+G +AY+L+LP+H ++HPVFHVSLLK A
Sbjct: 181 RRSLAKRINEKLSPRFYAPFQVFNKVGTIAYKLDLPSHIKIHPVFHVSLLKKA 339
Score = 97.1 bits (240), Expect(2) = 7e-59
Identities = 51/94 (54%), Positives = 63/94 (66%), Gaps = 2/94 (2%)
Frame = +3
Query: 1458 QPQPLPAALNEDHELLVEPESVLAVRETTMGQIEVLIQWHSLPACENSWESAAQLHEAFP 1517
Q QPLP L+ED +L +SVL RE G ++VLIQW +LP ENSWES A+L E F
Sbjct: 339 QSQPLPPMLSEDWKLQTYSDSVLDSRELQPGNVKVLIQWKNLPPSENSWESVAKLQEIFS 518
Query: 1518 DFPLEDKVKHLGGGID--EHRPSIKKVYERRNKR 1549
+ LEDKV LGGGID +H+P I KVY R++ R
Sbjct: 519 IYHLEDKVSLLGGGIDKHKHKPPIPKVYTRKHPR 620
>TC211627
Length = 1034
Score = 129 bits (325), Expect(2) = 3e-50
Identities = 86/213 (40%), Positives = 121/213 (56%), Gaps = 5/213 (2%)
Frame = +3
Query: 1326 LYGREPPVLLQGTTIPSKIAAVNDLQVGRDELLSDLRANLLKSQDMMRAYANKKRRDVDY 1385
+YG+ PP L + S + AV+ + + L L K QD M+ A+ RRD+ +
Sbjct: 243 MYGKPPPALPLYSAGTSTVEAVDAILHSLATIHHTLTCRLQKYQDSMKRIADSHRRDLTF 422
Query: 1386 QIGDEVFLKLQPYRRRSLAKKMNEKLSPRYYGPYPIVAKIGAVAYRLELPAHSRVHPVFH 1445
IGD V+++L PYR+ S+ KLS R+YGPY I A++G VAYRL+LP S++HP+FH
Sbjct: 423 NIGDWVYVRL*PYRQTSIQSTYT-KLSKRFYGPYQIQARVGQVAYRLQLPPTSKIHPIFH 599
Query: 1446 VSLLKTAVGTNFQPQ--PLPAALNEDHELLVEPESVL--AVRETTMGQI-EVLIQWHSLP 1500
VSLLK G P+ LP +H LV+P L + E+T I +VL+QW +L
Sbjct: 600 VSLLKVHHGP-IPPELLALPPFSTTNHP-LVQPLQFLDWKMDESTTPPIPQVLVQWTNLA 773
Query: 1501 ACENSWESAAQLHEAFPDFPLEDKVKHLGGGID 1533
+ +WES QL + + LEDKV GGID
Sbjct: 774 PEDTTWESWTQLKDI---YDLEDKVCFQTGGID 863
Score = 89.4 bits (220), Expect(2) = 3e-50
Identities = 40/79 (50%), Positives = 53/79 (66%)
Frame = +2
Query: 1246 LFLSSFWKELFRVSGTQLKFSSAYHPQTDGQTEVVNRCLEVYLRCLTGSRPRKWVTCLPW 1305
+F+S W ELF +SGT+L+FS+AYHPQTDGQTEV+NR LE YLR P+ W L
Sbjct: 2 IFISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSL 181
Query: 1306 AEFWFNSNYNRSAKMSPFQ 1324
AE +N++ + SPF+
Sbjct: 182 AE*CYNTSVHSGIGFSPFE 238
>BI425021
Length = 426
Score = 166 bits (419), Expect = 1e-40
Identities = 83/142 (58%), Positives = 101/142 (70%)
Frame = -1
Query: 1166 LQPLPVPTQVWTDVSMDFIGGLPKSNGKDTILVVVDRLTKYAHFIPLRHPFTAPEVAALF 1225
L PLPVP + W D+SMDFI GLP +G TI VVV+R +K H L TA VA+LF
Sbjct: 426 LCPLPVPQRPWEDLSMDFIVGLPPYHGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASLF 247
Query: 1226 LHEVVRLHGFPTTIVSDRDSLFLSSFWKELFRVSGTQLKFSSAYHPQTDGQTEVVNRCLE 1285
L+ V++LHGFP +IVSDRD LF+S FW++LFR+SGT L+ SSAYHPQTDGQTEV+NR +E
Sbjct: 246 LNIVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVIE 67
Query: 1286 VYLRCLTGSRPRKWVTCLPWAE 1307
YLR RPR +PW E
Sbjct: 66 QYLRAFVHGRPRNLGRFIPWVE 1
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 151 bits (382), Expect = 2e-36
Identities = 85/254 (33%), Positives = 138/254 (53%), Gaps = 19/254 (7%)
Frame = +1
Query: 644 IEKLVAEMLNAGIIRP-SISPYSSPVILVRKKDG------------------SWRFCVDY 684
+ K V ++L G+I P S S + SPV++V KK+G SW+ C+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 685 RALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGH 744
R LN+ T + FP+P +D++L+ + + +D GY+QI++ +D EK AF G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 745 YEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQL 804
+ + +PFGL NAP+TFQ M + ++ K VF DD ++ ++E + L VLQ
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 805 MDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWPVPKDLKSLRGFLGL 864
+ +L +N +K F + LGH +S + D K++ + P P ++K +R FLG
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 865 TGYYRRFVKDYGKI 878
+YRRF+KD+ K+
Sbjct: 721 ARFYRRFIKDFTKV 762
>AW570005
Length = 413
Score = 151 bits (381), Expect = 2e-36
Identities = 71/137 (51%), Positives = 93/137 (67%)
Frame = -2
Query: 850 PVPKDLKSLRGFLGLTGYYRRFVKDYGKIARPLTQLLKKDSFQWGPSPQQAFEALKHTLT 909
P P+ +SLRGFL LTG+YRRF+K Y +A PL+ LL KDSF W P AF+ALK+ +T
Sbjct: 412 PPPRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDSFVWSPEADVAFQALKNVVT 233
Query: 910 EIPTLAVPDFSKVFVLETDASGTGLGAVLTQEGKPLAFWSATLSDRSQAKSVYERELMAV 969
LA+PDF+K F +ETDASG+ +GAVL+QEG P+AF+S + S Y EL A+
Sbjct: 232 NTLVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAAI 53
Query: 970 VRAVQRWRHYLLGRHFI 986
V++WR YLLG H +
Sbjct: 52 TNVVKKWRQYLLGHHLV 2
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 145 bits (367), Expect(2) = 3e-36
Identities = 70/163 (42%), Positives = 104/163 (62%), Gaps = 1/163 (0%)
Frame = +1
Query: 850 PVPKDLKSLRGFLGLTGYYRRFVKDYGKIARPLTQLLKKD-SFQWGPSPQQAFEALKHTL 908
P K + +R F GL +YRRFV ++ +A PL +L+KK+ +F WG +QAF LK L
Sbjct: 97 PTLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKL 276
Query: 909 TEIPTLAVPDFSKVFVLETDASGTGLGAVLTQEGKPLAFWSATLSDRSQAKSVYERELMA 968
T+ P LA+PDFSK F LE DASG G+ AVL Q G P+A++S L + Y++EL A
Sbjct: 277 TKAPVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYA 456
Query: 969 VVRAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQFKWI 1011
++RA Q W H+L+ + F+I +D +SLK++ + + + KW+
Sbjct: 457 LIRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKWV 585
Score = 26.2 bits (56), Expect(2) = 3e-36
Identities = 11/29 (37%), Positives = 17/29 (57%)
Frame = +2
Query: 824 IEYLGHVLSAGTVAADPKKLEAMWLWPVP 852
I + G V+ V DP+K++A+ WP P
Sbjct: 17 IFFSGFVVGRNGVQMDPEKIKAIQEWPPP 103
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 147 bits (370), Expect = 5e-35
Identities = 86/254 (33%), Positives = 135/254 (52%), Gaps = 19/254 (7%)
Frame = +1
Query: 644 IEKLVAEMLNAGIIRP-SISPYSSPVILVRKKDGS------------------WRFCVDY 684
+ K V ++L AG+I P S S + SPV +V KK G WR C+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 685 RALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGH 744
R LN+ T + +P+P +D++L + + +D SGY+QI + +D EKTAF
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 745 YEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQL 804
+ + MPFGL NA +TFQ M + ++ KC VF DD + + + +L +VLQ
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 805 MDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWPVPKDLKSLRGFLGL 864
++ +L +N +K F + LGH +S + +KL+ + P P ++K + FLG
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 865 TGYYRRFVKDYGKI 878
G+YRRF+KD+ K+
Sbjct: 721 VGFYRRFIKDFTKV 762
>AW830191
Length = 372
Score = 144 bits (362), Expect = 4e-34
Identities = 67/123 (54%), Positives = 91/123 (73%)
Frame = +3
Query: 1288 LRCLTGSRPRKWVTCLPWAEFWFNSNYNRSAKMSPFQALYGREPPVLLQGTTIPSKIAAV 1347
LRCLTG++P++W L WAEFWFN+NYN S K++PF+ LYG +PP LL+G I S V
Sbjct: 6 LRCLTGTKPQQWPKRLSWAEFWFNTNYNNSLKLTPFKVLYGCDPPHLLKGAIISSTAEEV 185
Query: 1348 NDLQVGRDELLSDLRANLLKSQDMMRAYANKKRRDVDYQIGDEVFLKLQPYRRRSLAKKM 1407
N + RD++L DL+ NL K+Q+ M+ YA++ RR + +GD V+LKLQPYR+RSLA+K
Sbjct: 186 NVMTNDRDQMLHDLKGNLAKAQNQMK-YADRSRRSIPLNVGDWVYLKLQPYRQRSLARKT 362
Query: 1408 NEK 1410
NEK
Sbjct: 363 NEK 371
>TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, partial (7%)
Length = 810
Score = 127 bits (319), Expect = 4e-29
Identities = 72/157 (45%), Positives = 95/157 (59%), Gaps = 4/157 (2%)
Frame = +3
Query: 1372 MRAYANKKRRDVDYQIGDEVFLKLQPYRRRSLAKKMNEKLSPRYYGPYPIVAKIGAVAYR 1431
M+AYA++ RR V +GD V+LKLQPYR +SLAKK NEKLSPR+YGPY I +IG VA+
Sbjct: 3 MKAYADRSRRAVTLSVGDWVYLKLQPYRLKSLAKKRNEKLSPRFYGPYQIKKQIGLVAFE 182
Query: 1432 LELPAHSRVHPVFHVSLLKTAVGTNFQPQPLPAALNED-HELLVEPESVLAVRETTMGQI 1490
L+LP ++HPVFH SLLK AV PQPLP L+ED + +S L++ M +
Sbjct: 183 LDLPPARKIHPVFHASLLKKAVAATANPQPLPLMLSEDLSSEFFQLKSKLSI-TILMVLL 359
Query: 1491 EVLIQW---HSLPACENSWESAAQLHEAFPDFPLEDK 1524
+ W L N W+S+ +FP F L +
Sbjct: 360 KFSFNWKIFRILKPLGNPWKSS---RNSFPHFTLRTR 461
Score = 55.1 bits (131), Expect = 2e-07
Identities = 31/60 (51%), Positives = 34/60 (56%)
Frame = +2
Query: 1471 ELLVEPESVLAVRETTMGQIEVLIQWHSLPACENSWESAAQLHEAFPDFPLEDKVKHLGG 1530
EL V P V AV + G EVLIQ LP E +WES + E FP F LEDKV LGG
Sbjct: 299 ELRVFPAEVKAVHNNSNGIAEVLIQLEDLPDFEATWESVEVIKEQFPSFHLEDKVTLLGG 478
>CF922488
Length = 741
Score = 125 bits (313), Expect = 2e-28
Identities = 82/239 (34%), Positives = 126/239 (52%), Gaps = 1/239 (0%)
Frame = +3
Query: 671 VRKKDGSWRFCVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKA 730
V K+DG CVDYR LN + +KFP+P I+ L+D FS +D SGY+QI +
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 731 EDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLT 790
ED+EKT F T G + + M FGL N +T+Q M + ++ K V+ DD+++ S T
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 791 MEEHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWP 850
EEH+V+L ++ + + ++ L++N K +F + L + S + D K++ +
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 851 VPKDLKSLRGFLGLTGYYRRFVKDYGKIARPLTQLLKKDSF-QWGPSPQQAFEALKHTL 908
P K ++GFLG Y RF+ PL LL K+ F +W AFE +K L
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCL 719
>BQ627806
Length = 435
Score = 73.2 bits (178), Expect(2) = 8e-28
Identities = 37/84 (44%), Positives = 50/84 (59%)
Frame = +1
Query: 944 PLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQVV 1003
PL F + S Y REL A+ AV++WR YLLG HF+I TD +SLK L Q V
Sbjct: 181 PLPFSFMPFCSKLLRASTYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQ 360
Query: 1004 GEGQFKWISKLSGYDFEIQYKPGK 1027
Q ++++L G+D+ IQY+ GK
Sbjct: 361 TPEQQIYLARLMGFDYTIQYRAGK 432
Score = 70.9 bits (172), Expect(2) = 8e-28
Identities = 33/64 (51%), Positives = 42/64 (65%)
Frame = +3
Query: 885 LLKKDSFQWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETDASGTGLGAVLTQEGKP 944
LL KD F W +AF LK L + P L +PDF+ FV+ETDASG G+GA+L+Q P
Sbjct: 3 LLVKDQFHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHP 182
Query: 945 LAFW 948
LAF+
Sbjct: 183 LAFF 194
>BI317507
Length = 359
Score = 122 bits (306), Expect = 1e-27
Identities = 60/115 (52%), Positives = 80/115 (69%), Gaps = 1/115 (0%)
Frame = -1
Query: 781 FDDILIYSLTMEEHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADP 840
F +ILIYS + H++HLT VL ++ + L N KK F + IEYLGHV+S VA D
Sbjct: 359 FYNILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDS 180
Query: 841 KKLEAMWLWPVPKDLKSLRGFLGLTGYYRRFVKDYGKIA-RPLTQLLKKDSFQWG 894
K++++ WPVPK++K + FL LTGYYR+F+KDYGK+A RPLT L K D F+WG
Sbjct: 179 NKVKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLTKNDGFKWG 15
>CD487724
Length = 676
Score = 121 bits (304), Expect = 2e-27
Identities = 73/216 (33%), Positives = 111/216 (50%), Gaps = 2/216 (0%)
Frame = +3
Query: 439 IVVLVDCGATHNFISQSLAREMELQITETPAYTVEVGDGHVVKSKGVCKNLSVQIQGFVV 498
+V LVD G+THNF+ Q L ++ L TP V VG+GH +K +C+ + + IQ
Sbjct: 24 LVYLVDGGSTHNFVQQPLVSQLGLPCRSTPPLRVMVGNGHHLKCTTICEAIPISIQNIEF 203
Query: 499 TQDFFLFGLRGVDVVLGLEWLAGLGDIKANFEELTLKFKLGNKKVVLKGEPELLKKRASM 558
++ + G ++VLG++WL LG I ++ L+++F ++ V LKGE E +
Sbjct: 204 LVHLYVLPIVGANIVLGVQWLKTLGPILVDYNSLSMQFFYQHRLVQLKGESEAQLGLLNH 383
Query: 559 NALVKAIQQQGEGYVLQYQMLEKEVA--KAEETPELLKAVLSDFPALFAAPTELPPQRRH 616
+ L + Q +L + + + P+ ++ +L F ALF P LPP R
Sbjct: 384 HQLRRLHQTHEPVTYFHIAILTENTSPTSSPPLPQPIQHLLDQFSALFQ*PQGLPPARET 563
Query: 617 DHAIHLKEGASIPNIRPYRYPHYQKNEIEKLVAEML 652
DH IHL + N+R Y YPHY NEIE V ML
Sbjct: 564 DHHIHLLP*SEPVNMRLY*YPHY*NNEIEHQVNLML 671
>CA820403 weakly similar to GP|13273463|gb| pol protein integrase region
{Ginkgo biloba}, partial (52%)
Length = 421
Score = 95.9 bits (237), Expect(2) = 2e-25
Identities = 45/76 (59%), Positives = 59/76 (77%), Gaps = 1/76 (1%)
Frame = -3
Query: 1225 FLHEVVRLHGFPTTIVSDRDSLFLSS-FWKELFRVSGTQLKFSSAYHPQTDGQTEVVNRC 1283
F+ E V+LHG ++IVSD D LFL S FW ELF++ GT+LKFS AYHPQ D T+VVNRC
Sbjct: 335 FIKEAVKLHGCSSSIVSDWDRLFLIS*FWTELFKMEGTKLKFSLAYHPQPDSHTKVVNRC 156
Query: 1284 LEVYLRCLTGSRPRKW 1299
+E+ L+CLT S+ ++W
Sbjct: 155 IEMNLQCLTTSKRKQW 108
Score = 40.4 bits (93), Expect(2) = 2e-25
Identities = 17/34 (50%), Positives = 25/34 (73%)
Frame = -1
Query: 1196 ILVVVDRLTKYAHFIPLRHPFTAPEVAALFLHEV 1229
I+V+V R TKYAHF+ L HP+ A EV+ + L ++
Sbjct: 421 IMVIVYRFTKYAHFVVLSHPYLAKEVSEVLLKKL 320
>BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 424
Score = 107 bits (267), Expect = 4e-23
Identities = 51/120 (42%), Positives = 82/120 (67%)
Frame = +1
Query: 1365 LLKSQDMMRAYANKKRRDVDYQIGDEVFLKLQPYRRRSLAKKMNEKLSPRYYGPYPIVAK 1424
L ++Q +M +AN RR D +GD V+LK++P+R+ S+ +++ KL+ R+YGPY ++ +
Sbjct: 25 LERAQSLMVKHANNHRRPHDINVGDWVYLKIRPHRQGSMPPRLHPKLTARFYGPYLVMRQ 204
Query: 1425 IGAVAYRLELPAHSRVHPVFHVSLLKTAVGTNFQPQPLPAALNEDHELLVEPESVLAVRE 1484
+GAVA++L+LP+ +R+HPVFHVS LK A+G + + LP L EL P +L +RE
Sbjct: 205 VGAVAFQLQLPSEARIHPVFHVSQLKRALGNHQAQEELPPDLEHQAELYF-PVQILKIRE 381
>BI317638 weakly similar to GP|9294238|dbj| contains similarity to reverse
transcriptase~gene_id:K11J14.5 {Arabidopsis thaliana},
partial (5%)
Length = 420
Score = 92.4 bits (228), Expect(2) = 1e-21
Identities = 50/108 (46%), Positives = 63/108 (58%)
Frame = -2
Query: 1149 CDTCQRNKFDNLSPAGLLQPLPVPTQVWTDVSMDFIGGLPKSNGKDTILVVVDRLTKYAH 1208
C CQ K++ LL PL VP + W D+S+DFI GL + ILVVVD +K H
Sbjct: 326 CLDCQHTKYETKRIVDLLCPLLVPHRPWEDLSLDFITGLLPYHVHTAILVVVDHFSKGIH 147
Query: 1209 FIPLRHPFTAPEVAALFLHEVVRLHGFPTTIVSDRDSLFLSSFWKELF 1256
L TA VA LF+ V +LHG P ++VSD D LF+S FW+ELF
Sbjct: 146 LGMLPSSHTAHTVACLFIDSVAKLHGLPRSLVSDCDLLFVSHFWQELF 3
Score = 30.8 bits (68), Expect(2) = 1e-21
Identities = 11/26 (42%), Positives = 18/26 (68%)
Frame = -1
Query: 1117 GGHSGFFRTYRRLAAVVYWEGMKSDI 1142
GGH+G +T L+ +YW GM++D+
Sbjct: 411 GGHTGIAKTLA*LSKNIYWFGMRTDV 334
>TC233069
Length = 881
Score = 84.0 bits (206), Expect(2) = 6e-21
Identities = 39/75 (52%), Positives = 49/75 (65%)
Frame = +2
Query: 1138 MKSDIRDFVAECDTCQRNKFDNLSPAGLLQPLPVPTQVWTDVSMDFIGGLPKSNGKDTIL 1197
M D+ D + C CQ+NK+ GLLQPLP+P QVW D+SMDF+ LP S GK I
Sbjct: 224 MARDVCDHICACTNCQQNKYSTQK*FGLLQPLPIP*QVWKDISMDFVTHLPPS*GKKMIW 403
Query: 1198 VVVDRLTKYAHFIPL 1212
V+VD TKY+HF+ L
Sbjct: 404 VIVDCWTKYSHFLSL 448
Score = 37.0 bits (84), Expect(2) = 6e-21
Identities = 22/54 (40%), Positives = 26/54 (47%)
Frame = +1
Query: 1083 TLKDHRLFYKGKLVLSKQSSRIPILCKEFHASLLGGHSGFFRTYRRLAAVVYWE 1136
T +D L Y L + +S IL E H SLLGGHSG T RL W+
Sbjct: 61 TCQDGLLLYCTHLFIPLESELRAILLGECHDSLLGGHSGIKGTLNRLFVSFAWQ 222
>TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (5%)
Length = 761
Score = 99.0 bits (245), Expect = 1e-20
Identities = 62/153 (40%), Positives = 85/153 (55%), Gaps = 4/153 (2%)
Frame = +1
Query: 1387 IGDEVFLKLQPYRRRSLAKKMNEKLSPRYYGPYPIVAKIGAVAYRLELPAHSRVHPVFHV 1446
IGD V +KL+P+R+ S ++ KL+ RYYGP+ + ++G V YRL+L AHSR+HPVFHV
Sbjct: 4 IGDWVLVKLRPHRQGSASETTYSKLTKRYYGPFEVQERLGKVVYRLKLTAHSRIHPVFHV 183
Query: 1447 SLLKTAVG----TNFQPQPLPAALNEDHELLVEPESVLAVRETTMGQIEVLIQWHSLPAC 1502
SLLK VG T+ P P+ + L +S L + ++ VL+QW S
Sbjct: 184 SLLKAFVGDPETTHAGPLPVMRTEEATNTPLTVIDSKLVPADNGPRRM-VLVQWPSASRQ 360
Query: 1503 ENSWESAAQLHEAFPDFPLEDKVKHLGGGIDEH 1535
+ SWE L E + LEDKV G D H
Sbjct: 361 DASWEDWQVLRER---YNLEDKVLSEERGDDTH 450
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 62,988,701
Number of Sequences: 63676
Number of extensions: 861978
Number of successful extensions: 4948
Number of sequences better than 10.0: 138
Number of HSP's better than 10.0 without gapping: 4820
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4905
length of query: 1553
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1443
effective length of database: 5,635,272
effective search space: 8131697496
effective search space used: 8131697496
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0318.12


