

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.1
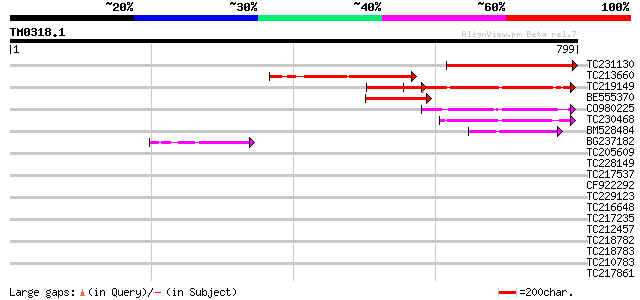
(799 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC231130 303 2e-82
TC213660 206 3e-53
TC219149 weakly similar to UP|SR40_YEAST (P32583) Suppressor pro... 199 5e-51
BE555370 189 5e-48
CO980225 145 5e-35
TC230468 similar to UP|Q9FHS2 (Q9FHS2) Gb|AAF03435.1, partial (11%) 130 2e-30
BM528484 117 2e-26
BG237182 100 2e-21
TC205609 40 0.003
TC228149 similar to UP|O02402 (O02402) Insoluble protein, partia... 39 0.009
TC217537 weakly similar to UP|Q8LK06 (Q8LK06) Methyl binding dom... 37 0.034
CF922292 37 0.045
TC229123 37 0.045
TC216648 weakly similar to UP|Q9FFQ8 (Q9FFQ8) Emb|CAB62360.1, pa... 37 0.045
TC217235 similar to UP|Q9LW88 (Q9LW88) Arabidopsis thaliana geno... 36 0.058
TC212457 UP|Q39871 (Q39871) Maturation polypeptide, complete 35 0.13
TC218782 similar to UP|Q41042 (Q41042) Pisum sativum L. (clone n... 34 0.29
TC218783 similar to UP|Q40363 (Q40363) NuM1 protein, partial (19%) 34 0.29
TC210783 weakly similar to UP|O14760 (O14760) Small intestinal m... 33 0.38
TC217861 similar to UP|Q761Y3 (Q761Y3) BRI1-KD interacting prote... 33 0.38
>TC231130
Length = 899
Score = 303 bits (777), Expect = 2e-82
Identities = 153/184 (83%), Positives = 163/184 (88%)
Frame = +3
Query: 616 DGEPKPFVLLNDLSDLLMLPKDMLIDRHVREEVCPSITLSLIIRVLCNFTPDEFCPDPVP 675
DGEPK FVLLNDLSDLLMLPKDMLIDR++R+EVCP+I LSLIIRVLCNFTPDEFCPDPVP
Sbjct: 3 DGEPKSFVLLNDLSDLLMLPKDMLIDRNIRQEVCPTIILSLIIRVLCNFTPDEFCPDPVP 182
Query: 676 GTVLEALNAETIAERRLSAESVRSFPYAAAAVVYVPPSSSNVAEKVAEAGGKSHLARNVS 735
GTVLEALNAETI ERRLSAES RSFPY A VVY+ PSS+NVAEKVAE GKSHLARNVS
Sbjct: 183 GTVLEALNAETIIERRLSAESARSFPYVAEPVVYMAPSSANVAEKVAETEGKSHLARNVS 362
Query: 736 AVQRRGYTSDEELEELDSPLTSIIDKLPLSPTVSANGQDNQKEHKSYTTTTNARYQLLRE 795
AVQRRGYTSDEELEELDSPLT IIDKLP SPTV+ NG+ N + + TTNARYQLLRE
Sbjct: 363 AVQRRGYTSDEELEELDSPLTFIIDKLPSSPTVTENGKGNNHKEQGGNPTTNARYQLLRE 542
Query: 796 VWSM 799
VWSM
Sbjct: 543 VWSM 554
>TC213660
Length = 582
Score = 206 bits (524), Expect = 3e-53
Identities = 112/208 (53%), Positives = 143/208 (67%), Gaps = 1/208 (0%)
Frame = +1
Query: 367 IVLREIISHAFGNSCQVSPLMRLAESNGAGNGKSASLKWKGFPNGKAGSGFTQFVEDWQE 426
IV +E+ + FG+ ++ +ES+G GN K + W++
Sbjct: 10 IVSKEVENIHFGDGPCIN-----SESDGTGNTLHKEEK----------DNTEKHFHRWED 144
Query: 427 TGTFTSALERVESWIFSRLVESVWWQALTPYMQSPAG-DFSSNKSFGRVLGPALGDHNQG 485
TF ALE+VE+WIFSR+VESVWWQ LTPYMQS A + SS K++ R +GD +QG
Sbjct: 145 PETFLVALEKVEAWIFSRIVESVWWQTLTPYMQSAAAKNSSSRKAYERRY--RVGDQDQG 318
Query: 486 NFSINLWRYAFEDAFQRLCPLRAGGHECGCLPVLARMVMEQCIARLDVAMFNAILRESAL 545
+FSI+LW+ AF+DA +R+CPLRAGGHECGCL V+AR+VMEQ ++RLDVAMFNAILRESA
Sbjct: 319 SFSIDLWKRAFKDACERICPLRAGGHECGCLLVIARLVMEQLVSRLDVAMFNAILRESAE 498
Query: 546 EIPTDPISDPILDSKVLPIPAGDLSFGS 573
E+P DPISDPI DS VLPIPAG FG+
Sbjct: 499 EMPMDPISDPISDSMVLPIPAGKSGFGA 582
>TC219149 weakly similar to UP|SR40_YEAST (P32583) Suppressor protein SRP40,
partial (8%)
Length = 1158
Score = 199 bits (505), Expect = 5e-51
Identities = 110/245 (44%), Positives = 155/245 (62%), Gaps = 2/245 (0%)
Frame = +1
Query: 555 PILDSKVLPIPAGDLSFGSGAQLKNSVGNWSRWLTDMFGMDVEDCVQEYQDSG--ENDER 612
P++ + P + FG+GAQLK ++GNWSRWLT +FGMD +D +++ D+ NDE
Sbjct: 163 PLVIPRFSPFHLVNQGFGAGAQLKTAIGNWSRWLTGLFGMDDDDPLEDIDDNDLDSNDES 342
Query: 613 QGGDGEPKPFVLLNDLSDLLMLPKDMLIDRHVREEVCPSITLSLIIRVLCNFTPDEFCPD 672
Q K F LLN LSDLLMLPKDML++ +R+EVCP + SLI ++L NF PDEFCPD
Sbjct: 343 QNTF---KSFHLLNALSDLLMLPKDMLLNASIRKEVCPMFSASLIKKILDNFVPDEFCPD 513
Query: 673 PVPGTVLEALNAETIAERRLSAESVRSFPYAAAAVVYVPPSSSNVAEKVAEAGGKSHLAR 732
P+P V EAL+++ E ES+ +FP AA Y PP ++ + E G +S L R
Sbjct: 514 PIPTDVFEALDSQDDLED--ENESISNFPCNAAPTAYSPPPAATITNITGEFGSESQLRR 687
Query: 733 NVSAVQRRGYTSDEELEELDSPLTSIIDKLPLSPTVSANGQDNQKEHKSYTTTTNARYQL 792
+ S+V R+ YTSD+EL+E++ PL+SI++ SP A+ + N K K + RY+L
Sbjct: 688 SKSSVVRKSYTSDDELDEINYPLSSILNSGSSSP---ASSKPNWK-WKDSRDESAVRYEL 855
Query: 793 LREVW 797
LR+VW
Sbjct: 856 LRDVW 870
Score = 112 bits (280), Expect = 6e-25
Identities = 55/91 (60%), Positives = 64/91 (69%), Gaps = 6/91 (6%)
Frame = +3
Query: 503 LCPLRAGGHECGCLPVLARMVMEQCIARLDVAMFNAILRESALEIPTDPISDPILDSKVL 562
LCP+RAGGHECG L VL +++MEQC+ARLDVAMFNAILRES +IPTDP+SDPI D KVL
Sbjct: 6 LCPIRAGGHECGFLSVLPKLIMEQCVARLDVAMFNAILRESDDDIPTDPVSDPISDPKVL 185
Query: 563 PIPAGDLSFGSGAQLKNS------VGNWSRW 587
PIP G KN + +WS W
Sbjct: 186 PIPPGQSRLWGWCTAKNCDW*LV*MADWSIW 278
>BE555370
Length = 280
Score = 189 bits (479), Expect = 5e-48
Identities = 89/93 (95%), Positives = 93/93 (99%)
Frame = +2
Query: 502 RLCPLRAGGHECGCLPVLARMVMEQCIARLDVAMFNAILRESALEIPTDPISDPILDSKV 561
RLCP+RAGGHECGCLPVLARMVMEQCIARLDVAMFNA+LRESALEIPTDPISDPI++SKV
Sbjct: 2 RLCPVRAGGHECGCLPVLARMVMEQCIARLDVAMFNALLRESALEIPTDPISDPIVNSKV 181
Query: 562 LPIPAGDLSFGSGAQLKNSVGNWSRWLTDMFGM 594
LPIPAGDLSFGSGAQLKNSVGNWSRWLTDMFGM
Sbjct: 182 LPIPAGDLSFGSGAQLKNSVGNWSRWLTDMFGM 280
>CO980225
Length = 806
Score = 145 bits (367), Expect = 5e-35
Identities = 92/221 (41%), Positives = 128/221 (57%), Gaps = 4/221 (1%)
Frame = -1
Query: 581 VGNWSRWLTDMFGMDVEDCVQEYQDSGENDERQGGDGEP--KPFVLLNDLSDLLMLPKDM 638
+G+WSRWL+D+F +D D + ++ E EP KPF LN LSDL+MLP DM
Sbjct: 692 IGDWSRWLSDLFSIDDSDSREVINENNEPK------CEPSFKPFQFLNALSDLMMLPLDM 531
Query: 639 LIDRHVREEVCPSITLSLIIRVLCNFTPDEFCPDPVPGTVLEALNAETIAERRLSAESVR 698
L D +R+EVCP +SL+ RV+ NF PDEF P P+P V EAL+ E I + ++
Sbjct: 530 LADGSMRKEVCPKFGISLMKRVVYNFVPDEFSPGPIPDAVFEALD-EDIED---DEGAIT 363
Query: 699 SFPYAAAAVVYVPPSSSNVAEKVAEAGGKSHLARNVSAVQRRGYTSDEELEELDSPLTSI 758
SFP +A + Y PP +S+V + E G K+ L R+ S V ++ YTSD+EL+ELDSPL+++
Sbjct: 362 SFPCSAGSTFYEPPPASSVVGMLQEVGTKTSL-RSGSFVLKKLYTSDDELDELDSPLSAL 186
Query: 759 --IDKLPLSPTVSANGQDNQKEHKSYTTTTNARYQLLREVW 797
D LS + G+ RY+LLRE W
Sbjct: 185 GMDDSSLLSKELVKGGR------------KVVRYELLREAW 99
>TC230468 similar to UP|Q9FHS2 (Q9FHS2) Gb|AAF03435.1, partial (11%)
Length = 707
Score = 130 bits (328), Expect = 2e-30
Identities = 83/193 (43%), Positives = 112/193 (58%), Gaps = 1/193 (0%)
Frame = +1
Query: 606 SGENDERQGGDGEPKPFVLLNDLSDLLMLPKDMLIDRHVREEVCPSITLSLIIRVLCNFT 665
S EN+E + + KPF LN LSDL+MLP DML D + +EVCP +SLI RV+ NF
Sbjct: 7 SXENNESKX-ESSFKPFQFLNALSDLMMLPLDMLADGSMIKEVCPKFGISLIKRVVYNFV 183
Query: 666 PDEFCPDPVPGTVLEALNAETIAERRLSAESVRSFPYAAAAVVYVPPSSSNVAEKVAEAG 725
PDEF P P+P V +AL+ E I + ++ SFP A +Y PP +S+V K+ E G
Sbjct: 184 PDEFSPGPIPDAVYDALDNEDIQD---GEGAITSFPCPAGFTLYAPPPASSVVGKLQEVG 354
Query: 726 GKSHLARNVSAVQRRGYTSDEELEELDSPLTSI-IDKLPLSPTVSANGQDNQKEHKSYTT 784
K+ L R S V ++ YTSD+EL+ELDSPL+++ +D LS +K
Sbjct: 355 NKTSL-RTGSFVLKKLYTSDDELDELDSPLSALGMDDSSLS--------SKEKLALVKGG 507
Query: 785 TTNARYQLLREVW 797
RY+LLRE W
Sbjct: 508 RKVVRYELLREAW 546
>BM528484
Length = 427
Score = 117 bits (294), Expect = 2e-26
Identities = 63/132 (47%), Positives = 80/132 (59%)
Frame = +3
Query: 647 EVCPSITLSLIIRVLCNFTPDEFCPDPVPGTVLEALNAETIAERRLSAESVRSFPYAAAA 706
EVCP +LI ++L NF PDE CPDPVP V EALN+E E E V +FP AA
Sbjct: 6 EVCPMFNATLIKKILDNFVPDELCPDPVPSNVFEALNSEN--EMEDGKEYVNNFPCIAAP 179
Query: 707 VVYVPPSSSNVAEKVAEAGGKSHLARNVSAVQRRGYTSDEELEELDSPLTSIIDKLPLSP 766
+ Y PP ++++A V E G KS L RN S+V R+ +TSD+EL+EL SPL+SI + SP
Sbjct: 180 IAYSPPPATSIASIVGEIGSKSQLRRNKSSVVRKSHTSDDELDELKSPLSSIFFSVSSSP 359
Query: 767 TVSANGQDNQKE 778
V KE
Sbjct: 360 KVLTKSSLKFKE 395
>BG237182
Length = 422
Score = 100 bits (250), Expect = 2e-21
Identities = 64/149 (42%), Positives = 86/149 (56%)
Frame = +2
Query: 197 KNTSSNNSKPVKAPVKASSESSEGVDENHVLEVKEIEILDGASNGAQSLGSEDERHETVN 256
K+ N+K V+ P S + ++EN +L G + + S D R +
Sbjct: 20 KSERLKNTKSVRLP--GDSVRNAELNENGIL---------GDAQNSSGNRSNDRRDSKIL 166
Query: 257 AEENGEHEDKAALELKIEEMELRVEKLEEELREVAALEVSLYSVVPEHGSSAHKVHTPAR 316
A+E L+ KIE +E +++ LE ELRE AA+E +LY+VV EHG+S KVH PAR
Sbjct: 167 AKEIRS----GTLDGKIEHLEKKIKMLEGELREAAAIEAALYTVVAEHGNSTSKVHAPAR 334
Query: 317 RLSRLYLHASKHWTQNRRATIAKNTVSGL 345
RLSRLYLHASK Q RRA AK++VSGL
Sbjct: 335 RLSRLYLHASKENLQERRAGAAKSSVSGL 421
>TC205609
Length = 1142
Score = 40.4 bits (93), Expect = 0.003
Identities = 44/167 (26%), Positives = 68/167 (40%), Gaps = 2/167 (1%)
Frame = +2
Query: 7 RKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTASEN 66
+KA +++KGS+ + EDK+ SK K SK+ +N + +++T S+
Sbjct: 119 QKAKDGNKSKGSKANSKSEDKV--SRKSKDGTPKSGSSKSIVAAKKMSNKLKNTDT-SKT 289
Query: 67 SETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKE-GNGEVSDTDTVKDSV 125
SE+ ++ E L K A K K G +V+ T VK +
Sbjct: 290 SESKDDDSSKPKPSAKSKHETLKSGKSKQETPKAAISKGKPVKSGGKTDVNGTGKVKSGL 469
Query: 126 SSQGDSFTNEDEKVEKAS-KDAKSKVKVSPSESSRGLKERSDRKTNK 171
+ F NE+ V +D+K K S LK R RKT K
Sbjct: 470 LKR-KHFENENSDVSAGEIEDSKGKTASSSKAQGSELKSRKRRKT*K 607
>TC228149 similar to UP|O02402 (O02402) Insoluble protein, partial (4%)
Length = 1008
Score = 38.9 bits (89), Expect = 0.009
Identities = 54/219 (24%), Positives = 77/219 (34%), Gaps = 5/219 (2%)
Frame = -2
Query: 3 ETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNT 62
E E KA T R+ RR +K NSS N+K + TP+ S+ N S T
Sbjct: 836 EEELPKAKEKPVT--GARSLRRSNKNLNSNSSNNNNKKPSSISTPKAGSSWNK---PSET 672
Query: 63 ASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVK 122
E D RS A ++ + +AS G G +
Sbjct: 671 VKPEKSKAEGGA-----DKKRSAAADFLKRIKRNTSAEASKGGSGGGSGGGGGGGGGSSS 507
Query: 123 DSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNK-----LQSKVS 177
S G + E +K+ K K K + S + G +D++ NK QSK S
Sbjct: 506 SSKGGGGGNGAKEQKKMVNNGKGDKGKERASRHNNVGGGSGSADKRNNKNIENSSQSKRS 327
Query: 178 DSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSE 216
K T S + S++ K K P K S +
Sbjct: 326 VGRPPKKAAETNAGSAKRGRENSASAGKD-KRPKKRSKK 213
>TC217537 weakly similar to UP|Q8LK06 (Q8LK06) Methyl binding domain protein
MBD109, partial (13%)
Length = 968
Score = 37.0 bits (84), Expect = 0.034
Identities = 52/244 (21%), Positives = 92/244 (37%), Gaps = 4/244 (1%)
Frame = +1
Query: 17 GSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTASENSETYENVVID 76
G+ T RR ++ S K TES+ P+ R+ ++ + E E + +
Sbjct: 55 GTGETPRRSTRI----SEKAKATPPTESEPPKKRTKRSSASQKETSQEEKEEETKEAEMQ 222
Query: 77 YVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDSVSSQGDSFTNED 136
VDD + + + + K D E ++G V DTD K + S + +
Sbjct: 223 EVDDTTKDDNDIEKEK----------DVVMENQDGK-SVEDTDVNKSTHSGEAKA----G 357
Query: 137 EKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQKKPMNSTKGPSRV-- 194
E VE ++ KS + + L D+ +K+ + + ++P TK R
Sbjct: 358 EYVEVPIEEEKS------NAADGELPALKDKVDDKVFLRKDEEKIEQPQEETKEYRRTGV 519
Query: 195 -TNKNTSSNNSKPVKAPVKASSESSEGVD-ENHVLEVKEIEILDGASNGAQSLGSEDERH 252
T + K V+ EGV+ E E+ I++G + + + E
Sbjct: 520 PEKSETCTTADKTVEV---------EGVNKEVRSTRQFEVGIIEGTKVNSDQINKKAEAE 672
Query: 253 ETVN 256
TVN
Sbjct: 673 LTVN 684
>CF922292
Length = 722
Score = 36.6 bits (83), Expect = 0.045
Identities = 32/145 (22%), Positives = 65/145 (44%)
Frame = -2
Query: 1 MKETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDS 60
+KE ++ K+ SQ K S T+ +N +++EK ++S Q +S DS
Sbjct: 328 VKEKQEEKSDEKSQEKPSEDTKT-------ENQDTSVSEKRSDSDESQQKS-------DS 191
Query: 61 NTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDT 120
+ + + S++ E+ E K ++ + + SD+++ +K+ SD+D
Sbjct: 190 DESQQKSDSDES-----------------EKKSDSAESEKKSDSDESEKK-----SDSDE 77
Query: 121 VKDSVSSQGDSFTNEDEKVEKASKD 145
+ S S + + DE+ K+ D
Sbjct: 76 TEKSSESNDNKQFDSDERENKSDSD 2
>TC229123
Length = 1034
Score = 36.6 bits (83), Expect = 0.045
Identities = 40/161 (24%), Positives = 68/161 (41%)
Frame = +3
Query: 18 SRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVGDSNTASENSETYENVVIDY 77
SR++ ED Q SSK + K S ++ NN +SEN + + + +Y
Sbjct: 72 SRKSSNGED---QGTSSKDTDLKKKGSTXRGNKPTRNNTAEKEKYSSENKDEFSRLKGEY 242
Query: 78 VDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDSVSSQGDSFTNEDE 137
+ LA K + + A EK+ + + D D ++
Sbjct: 243 --------DKLALEKYSEVTTLLA---EKKFMWNQYNIMEND--------YADKLRTKEA 365
Query: 138 KVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSD 178
+VEKA++ K K+ VS E + K D K ++LQSK+++
Sbjct: 366 EVEKANE--KIKILVSSMEQLQSEKYEKDSKISELQSKMAE 482
>TC216648 weakly similar to UP|Q9FFQ8 (Q9FFQ8) Emb|CAB62360.1, partial (41%)
Length = 2064
Score = 36.6 bits (83), Expect = 0.045
Identities = 49/224 (21%), Positives = 84/224 (36%), Gaps = 13/224 (5%)
Frame = +1
Query: 2 KETEKRKASRNS---QTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQDRSAANNLVG 58
KE + +K +R S ++ G TE K Q + S +K + D++ ++ G
Sbjct: 922 KEQKGKKRTRKSVPSKSPGEASTETPAKKQKQTSQSGK-KQKQSSDDDEDDKAELSDAKG 1098
Query: 59 ----DSNTASENSETYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGE 114
D + A N+E+ D+ +RSEE + K + +A K
Sbjct: 1099 VSQEDEDVAVPNNESD--------DEESRSEEEEEKSKSRKRTSKKAVKESSVSKADRTS 1254
Query: 115 VSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQS 174
VKD+ S +EK K SK V+ +S+ +S + +K Q
Sbjct: 1255 SVKKTPVKDAKS------------IEKTKKKPTSKKGVAEHDSASASVSKSKQPASKKQK 1398
Query: 175 KVSDSNQKKPMNSTK------GPSRVTNKNTSSNNSKPVKAPVK 212
S+ K ++K + V ++ S +N K P K
Sbjct: 1399 TASEKQDTKGKAASKKRTDKSSKALVKDQGKSKSNKKAKAEPTK 1530
>TC217235 similar to UP|Q9LW88 (Q9LW88) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MSL1, partial (36%)
Length = 1253
Score = 36.2 bits (82), Expect = 0.058
Identities = 32/155 (20%), Positives = 61/155 (38%), Gaps = 16/155 (10%)
Frame = +1
Query: 107 EQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSD 166
++ + N D++ D + Q +S TNE E SKDA + ++ +
Sbjct: 1 DESKSNDTAKDSEKAGDGEAKQPESKTNEAEDKLTESKDAAEESNADKEHTAEKESTDDE 180
Query: 167 RKTNKLQSK---VSDSNQKKPMNSTKGPSRV-------------TNKNTSSNNSKPVKAP 210
K +K Q+K S + KK + S++ +KN ++NN +
Sbjct: 181 SKVDKEQNKDVTESGNEDKKDATHNESASKLDKEKTGDGKDSENDDKNENTNNVDKKDSK 360
Query: 211 VKASSESSEGVDENHVLEVKEIEILDGASNGAQSL 245
+++ S+EG +K ++L + N L
Sbjct: 361 AESAEPSAEGG------HLKSFQLLSSSQNAFTGL 447
>TC212457 UP|Q39871 (Q39871) Maturation polypeptide, complete
Length = 1741
Score = 35.0 bits (79), Expect = 0.13
Identities = 70/324 (21%), Positives = 123/324 (37%), Gaps = 33/324 (10%)
Frame = +1
Query: 2 KETEKRKASRNSQTKGSRRTERREDKLHQDNSSKTLNEKGTESKTPQD--RSAANNLVGD 59
KE E+ R + G ++++ HQ++ K G+ K QD +A +VG
Sbjct: 112 KELEQVNRERRDRDFGVVAEQQQQH--HQEDQQKR-GVIGSMFKAVQDTYENAKEAVVGK 282
Query: 60 ---SNTASENSETYENVVIDYVDDVNRSEEA--LAEMKVNAMVANQASDTEKEQKEGNGE 114
+N A N+E +V I DDV+ + E ++ K + + + + + GE
Sbjct: 283 KEATNNAYSNTEVIHDVNIQ-PDDVSATGEVRDISATKTHDIYDSATDNNNNKTGSKVGE 459
Query: 115 VSD-----TDTVKDSVSSQGDSFTN-EDEKVEKASKDAKSKVKVSPSESSRGLKERSDRK 168
+D KD+ + +T+ +K ++A K K S+ KER D
Sbjct: 460 YADYASQKAKETKDATMEKAGEYTDYASQKAKEAKKTTMEKGGEYKDYSAEKAKERKDAT 639
Query: 169 TNKLQSKVSDSNQKKPMNSTKGPSRVTNK----------NTSSNNSKPVKAPVKASSESS 218
NK+ + +K + +G NK T V + ++
Sbjct: 640 VNKMGEYKDYAAEK----AKEGKDATVNKMGEYKDYAAEKTKEGKDATVNKMGEYKDYTA 807
Query: 219 EGVDENHVLEVKEI-EILDGASNGAQSL-----GSEDE----RHETVNAEENGEHEDKAA 268
E E + ++ E+ D AS+ A+ G ++E ET A N E K A
Sbjct: 808 EKAKEGKDTTLGKLGELKDTASDAAKRAVGYLSGKKEETKEMASETAEATANKAGEMKEA 987
Query: 269 LELKIEEMELRVEKLEEELREVAA 292
+ K E + E+++ AA
Sbjct: 988 TKKKTAETAEAAKNKAGEIKDRAA 1059
>TC218782 similar to UP|Q41042 (Q41042) Pisum sativum L. (clone na-481-5),
partial (20%)
Length = 925
Score = 33.9 bits (76), Expect = 0.29
Identities = 25/119 (21%), Positives = 51/119 (42%)
Frame = +1
Query: 106 KEQKEGNGEVSDTDTVKDSVSSQGDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERS 165
K+ NG +S + SS S ++ED D+ + +V+ + ++ +K +
Sbjct: 10 KKTPTKNGNLSTPAKKGKAASSSSSSDSSED--------DSSDEDEVATKKQTKEVKVQK 165
Query: 166 DRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDEN 224
++ + S+S +KP P + + + ++ K ASS SSE D++
Sbjct: 166 GKEESSSDDSSSESEDEKPAAKVAVPPKNQSAKNGTLSTPAEKGKPAASSSSSESSDDD 342
>TC218783 similar to UP|Q40363 (Q40363) NuM1 protein, partial (19%)
Length = 870
Score = 33.9 bits (76), Expect = 0.29
Identities = 49/210 (23%), Positives = 88/210 (41%), Gaps = 4/210 (1%)
Frame = +3
Query: 12 NSQTKGSRRTERRE-DKLHQDNSSKT--LNEKGTESKTPQDRSAANNLVGDSNTASENSE 68
++ TK ++ +E + D D+SS +K T +K P A V S+ S S
Sbjct: 291 HASTKKTQPSESSDSDSSDSDSSSDEGKSKKKPTTAKLPTLPVAPAKKVESSDDESSESS 470
Query: 69 TYENVVIDYVDDVNRSEEALAEMKVNAMVANQASDTEKEQKEGNGEVSDTDTVKDSVSSQ 128
+N V V++ A A+ KV + + + D+ ++ +G ++ D D D
Sbjct: 471 DEDNDAKPAVTAVSKPS-ARAQKKVES---SDSDDSSSDEDKGKMDIDDDDDSSDE---- 626
Query: 129 GDSFTNEDEKVEKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQKKPMNST 188
+E+ + +KA K++K ESS +E S+ D +++KP +
Sbjct: 627 -----SEEPQKKKAVKNSK--------ESSDSSEEDSE-----------DESEEKPSKTP 734
Query: 189 KGPSR-VTNKNTSSNNSKPVKAPVKASSES 217
+ R V + + + K K PV ES
Sbjct: 735 QKRGRDVEMVDAALSEKKAPKTPVTPREES 824
Score = 30.8 bits (68), Expect = 2.5
Identities = 25/85 (29%), Positives = 39/85 (45%)
Frame = +3
Query: 140 EKASKDAKSKVKVSPSESSRGLKERSDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNT 199
E A+K ++VKV +G +E S ++ SDS +KP PS+ +
Sbjct: 12 ELATKKQMNEVKVQ-----KGKEESSSDDSS------SDSEDEKPAAKVAVPSKNQSAKN 158
Query: 200 SSNNSKPVKAPVKASSESSEGVDEN 224
+ ++ K ASS SSE D+N
Sbjct: 159 GTLSTLAKKGKPAASSSSSESSDDN 233
>TC210783 weakly similar to UP|O14760 (O14760) Small intestinal mucin MUC3
(Fragment), partial (5%)
Length = 735
Score = 33.5 bits (75), Expect = 0.38
Identities = 37/139 (26%), Positives = 58/139 (41%), Gaps = 9/139 (6%)
Frame = +3
Query: 108 QKEGNGEVSDTDTVKDSVSSQGDSFTNE--DEKVEKASKDAKS-KVKVSPSESSRGLKER 164
++E EVS TD S G SF + DE EK + KV + +KE
Sbjct: 210 EEESVNEVSKTD-------SNGKSFQEKTLDEITEKKDLETDGGKVLENNGIKKNIVKEI 368
Query: 165 SDRKTNKLQSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASS------ESS 218
D+K ++ KV + + + K + + + K V VK + E+
Sbjct: 369 EDKKAVGVE-KVEEDKKNGVVEDAKEDKKEDGVKEAKEDKKEVVEEVKEDNKEDGVEEAK 545
Query: 219 EGVDENHVLEVKEIEILDG 237
E E+ V EVKE++ +DG
Sbjct: 546 ENKKEDGVEEVKEVKEVDG 602
>TC217861 similar to UP|Q761Y3 (Q761Y3) BRI1-KD interacting protein 132
(Fragment), partial (27%)
Length = 1293
Score = 33.5 bits (75), Expect = 0.38
Identities = 34/143 (23%), Positives = 58/143 (39%), Gaps = 29/143 (20%)
Frame = +1
Query: 173 QSKVSDSNQKKPMNSTKGPSRVTNKNTSSNNSKPVKAPVKASSESSEGVDENH---VLEV 229
+S ++D N++ +K NK + K K S + V++NH V V
Sbjct: 328 KSNINDQNEQVAGGHSK------NKEEGKSKEKKKKKSKLVSESLATNVEDNHLESVATV 489
Query: 230 KEIEILDGASNGAQSL-GSEDER-------------------------HETVNAEENGEH 263
+E ++ DG S A+ + GSE E+ +ETV+ EEN E
Sbjct: 490 EENKVKDGVSTDAKVINGSEKEKKSKSKMKKKDKQSGQGDVIEQIGDPNETVSKEENIEA 669
Query: 264 EDKAALELKIEEMELRVEKLEEE 286
+K + ++ + R + EE
Sbjct: 670 SNKEMMHEVEKDSKKRKRPISEE 738
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.127 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,884,259
Number of Sequences: 63676
Number of extensions: 391804
Number of successful extensions: 1883
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 1845
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1870
length of query: 799
length of database: 12,639,632
effective HSP length: 105
effective length of query: 694
effective length of database: 5,953,652
effective search space: 4131834488
effective search space used: 4131834488
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0318.1


