

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
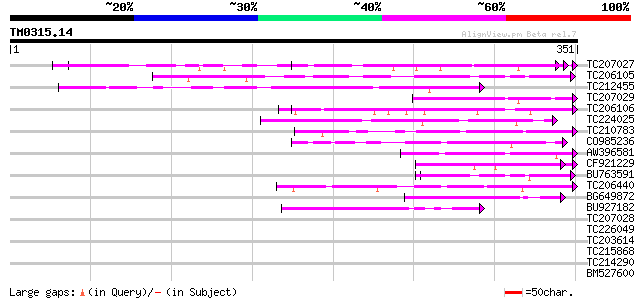
Query= TM0315.14
(351 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 60 1e-09
TC206105 weakly similar to UP|Q7RH28 (Q7RH28) Mature-parasite-in... 55 6e-08
TC212455 weakly similar to UP|Q7QIA8 (Q7QIA8) AgCP3984 (Fragment... 53 2e-07
TC207029 weakly similar to UP|Q7R8E7 (Q7R8E7) Ran-binding protei... 50 2e-06
TC206106 weakly similar to UP|Q7RK82 (Q7RK82) Maebl (Fragment), ... 50 2e-06
TC224025 weakly similar to UP|Q6HXC4 (Q6HXC4) Conserved domain p... 49 3e-06
TC210783 weakly similar to UP|O14760 (O14760) Small intestinal m... 47 1e-05
CO985236 45 5e-05
AW396581 45 5e-05
CF921229 45 6e-05
BU763591 44 1e-04
TC206440 43 2e-04
BG649872 42 4e-04
BU927182 42 5e-04
TC207028 similar to UP|Q9LW95 (Q9LW95) KED, partial (13%) 41 0.001
TC226049 similar to UP|O24078 (O24078) Protein phosphatase 2C, p... 40 0.001
TC203614 UP|Q94LX2 (Q94LX2) Beta-conglycinin alpha subunit, comp... 40 0.001
TC215868 similar to UP|Q9LJ56 (Q9LJ56) Gb|AAD43149.1, partial (50%) 39 0.003
TC214290 homologue to UP|Q6UJX5 (Q6UJX5) Molecular chaperone Hsp... 39 0.003
BM527600 similar to PIR|T05883|T058 ATP-dependent helicase F6H11... 39 0.004
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 60.5 bits (145), Expect = 1e-09
Identities = 86/323 (26%), Positives = 146/323 (44%), Gaps = 8/323 (2%)
Frame = +3
Query: 37 DGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMVRKCRRAGLDGDGEKEREA 96
DGEE +EK K +EK KK E +K D + +K++ G + DGE + E
Sbjct: 9 DGEEKKEKDK--KEK-----KKKENKDKGDKTDVEKVK----------GKENDGEDDEEK 137
Query: 97 GEDLESGRRDGLDGYGEEER-----EIGEVDEEKEVKLGK--KFGMAWIQVNHYELREHG 149
E + ++D DG + E+ + G+ DEEK+ K K K ++ + ++ G
Sbjct: 138 KEKKKKEKKDK-DGKTDAEKIKKKEDDGKDDEEKKEKKKKYDKIDAXKVKGKEDDGKDEG 314
Query: 150 ERRKCGLLEFNFSIDLDSGIRDEIDEDGEEEREADGLHVDEENEREVDDGNEETELKYDK 209
+ K D + G D DGEE++E ++E ++E D +EET+
Sbjct: 315 NKEKK---------DKEKG-----DGDGEEKKEKKDK--EKEKKKEKKDKDEETD----- 431
Query: 210 GYEMVRILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLH 269
LKE+ + ++ N K + ++E+E++ + +KD+
Sbjct: 432 ------------TLKEKGKNDEGEDDEGNKKK--------KKDKKEKEKDHKKEKKDKEE 551
Query: 270 GEEEEER-EVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHV 328
GE+E+ + EVSV + + E I +E E E++ ++G +E V
Sbjct: 552 GEKEDSKVEVSVRDIDIE---------EIKKEGEK---------EDKGKDGGKE-----V 662
Query: 329 KEKKKIEKLEKENEEKKVKRGKD 351
KEKKK E +K+ ++KKV GKD
Sbjct: 663 KEKKKKEDKDKKEKKKKV-TGKD 728
Score = 58.9 bits (141), Expect = 3e-09
Identities = 76/317 (23%), Positives = 144/317 (44%), Gaps = 2/317 (0%)
Frame = +3
Query: 27 ESGLRDRRDGDGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMVRKCRRAGL 86
E ++++D DG+ D EK+K E+ D +K E K D + K++ G
Sbjct: 141 EKKKKEKKDKDGKTDAEKIKKKEDDGKDDEEKKEKKKKYDKIDAXKVK----------GK 290
Query: 87 DGDGEKE-REAGEDLESGRRDGLDGYGEEEREIGEVDEEKEVKLGKKFGMAWIQVNHYEL 145
+ DG+ E + +D E G DG GEE++E + D+EKE K KK +
Sbjct: 291 EDDGKDEGNKEKKDKEKG-----DGDGEEKKE--KKDKEKEKKKEKK-----------DK 416
Query: 146 REHGERRKCGLLEFNFSIDLDSGIRDE-IDEDGEEEREADGLHVDEENEREVDDGNEETE 204
E + K + G DE D++G ++++ D ++++++E D EE E
Sbjct: 417 DEETDTLK------------EKGKNDEGEDDEGNKKKKKDKKEKEKDHKKEKKD-KEEGE 557
Query: 205 LKYDKGYEMVRILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVE 264
+ K VR + + KE EK K DG V+E++++E +
Sbjct: 558 KEDSKVEVSVRDIDIEEIKKE-------GEKEDKGK------DGGKEVKEKKKKE----D 686
Query: 265 KDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDEEEEREGDEEKE 324
KD+ +E++++V+ ++ K+L K+ I+ +++ + E++ D E++
Sbjct: 687 KDK----KEKKKKVTGKDKTKDLSTLKQKLEKINGKIQPLL----------EKKADIERQ 824
Query: 325 VKHVKEKKKIEKLEKEN 341
+K V+ + + E +N
Sbjct: 825 IKEVEAEGHVVNEENKN 875
Score = 53.1 bits (126), Expect = 2e-07
Identities = 53/202 (26%), Positives = 96/202 (47%), Gaps = 30/202 (14%)
Frame = +3
Query: 175 EDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYKQE 234
+DGEE++E D E+ ++E D ++T+++ KG E N E+ +K K+E
Sbjct: 6 DDGEEKKEKDK---KEKKKKENKDKGDKTDVEKVKGKE------NDGEDDEEKKEKKKKE 158
Query: 235 KR---------------------YNSKNLIVEYDGLD----RVEEEEEREVGEVEK---D 266
K+ K +YD +D + +E++ ++ G EK +
Sbjct: 159 KKDKDGKTDAEKIKKKEDDGKDDEEKKEKKKKYDKIDAXKVKGKEDDGKDEGNKEKKDKE 338
Query: 267 RLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDE--EEEREGDEEKE 324
+ G+ EE++E +EK+ K K+ + E G D + DE +++++ +EKE
Sbjct: 339 KGDGDGEEKKEKKDKEKEKK-KEKKDKDEETDTLKEKGKNDEGEDDEGNKKKKKDKKEKE 515
Query: 325 VKHVKEKKKIEKLEKENEEKKV 346
H KEKK E+ EKE+ + +V
Sbjct: 516 KDHKKEKKDKEEGEKEDSKVEV 581
>TC206105 weakly similar to UP|Q7RH28 (Q7RH28) Mature-parasite-infected
erythrocyte surface antigen, partial (5%)
Length = 858
Score = 54.7 bits (130), Expect = 6e-08
Identities = 64/270 (23%), Positives = 126/270 (45%), Gaps = 8/270 (2%)
Frame = +1
Query: 89 DGEKEREAGEDLESGRRDGLD---GYGEEEREIGEVDEEKEVKLGKKFGMAWIQVNHYEL 145
+ E E E +SG D + + EE +G++ E ++ L + + +N++EL
Sbjct: 172 EAEAENIEAEKAQSGVEDKISQSVSFKEETNVVGDLPEAQKKALDELKKLVQEALNNHEL 351
Query: 146 ---REHGERRKCGLLEFNFSIDLDSGIRDEID-EDGEEEREADGLHVDEENEREVDDGNE 201
+ E++K + ++E++ +G++E E ++E+ E EV + +
Sbjct: 352 TAPKPEPEKKKPAAEK-----------KEEVEVTEGKKEAEV----IEEKKEVEVTEEKK 486
Query: 202 ETELKYDKGYEMVRILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDGLDRVEEEEEREVG 261
E E+ +K V++E+ + +EK+ +E EE++
Sbjct: 487 EIEVTEEKK--------EAEVIEEKKEVEVTEEKK--------------EIEVTEEKKEA 600
Query: 262 EVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDEEEEREGDE 321
EV++++ GE EE++ V EEK KE E+ + + E + + + E E EG
Sbjct: 601 EVKEEKKEGEVTEEKKEVEVTEEK-----KEAEVIVEEKKEVEVTE--EKKEVEVTEG-- 753
Query: 322 EKEVKHVKEKKKIEKLEKENE-EKKVKRGK 350
+KEV+ ++EKK+ E E++ E E +V+ K
Sbjct: 754 KKEVEVIEEKKETEVTEEKKEVEVEVREEK 843
Score = 30.4 bits (67), Expect = 1.3
Identities = 30/124 (24%), Positives = 53/124 (42%), Gaps = 27/124 (21%)
Frame = +1
Query: 255 EEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDG--- 311
E E E E EK + E++ + VS E + E + EL+ +++ L+
Sbjct: 172 EAEAENIEAEKAQSGVEDKISQSVSFKEETNVVGDLPEAQKKALDELKKLVQEALNNHEL 351
Query: 312 -----------------DEEEEREG-------DEEKEVKHVKEKKKIEKLEKENEEKKVK 347
+E E EG +E+KEV+ +EKK+IE E++ E + ++
Sbjct: 352 TAPKPEPEKKKPAAEKKEEVEVTEGKKEAEVIEEKKEVEVTEEKKEIEVTEEKKEAEVIE 531
Query: 348 RGKD 351
K+
Sbjct: 532 EKKE 543
>TC212455 weakly similar to UP|Q7QIA8 (Q7QIA8) AgCP3984 (Fragment), partial
(11%)
Length = 944
Score = 52.8 bits (125), Expect = 2e-07
Identities = 63/273 (23%), Positives = 113/273 (41%), Gaps = 9/273 (3%)
Frame = +1
Query: 31 RDR-RDGDGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMVRKCRRAGLDGD 89
RDR R D + DR+ + +++E GK++E W KER++ RE+ R D
Sbjct: 250 RDRDRSRDRDVDRDLHREKKQEETSRGKEIE-WEHKRGKERERSRERYRR---------D 399
Query: 90 GEKEREAGEDLESGRRDGLDGYGEEEREIGEVDEEKEVKLGKKFGMAWIQVNHYELREHG 149
EK+R E E G+ +EKE +W V + R
Sbjct: 400 AEKDRS-----------------REREEDGDRRQEKE------RDRSWDTVFERDRRREK 510
Query: 150 ERRKCGLLEFNFSIDLDSGIRDEIDEDGEEEREADGLHVDEENEREVDDGN-EETELKYD 208
ER + S D G + + D + E + + D ++ D N ++ L+++
Sbjct: 511 ERDR--------SRDRTIGGKRDRDPENERDDKNRERRDDRYGHKDRDTANGKDRHLRHE 666
Query: 209 KGYEMVRILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDGLDRVEEEEER-------EVG 261
G + H+ +E ++Y E++ NS N + Y+ + E +E E
Sbjct: 667 DGNDNGDRYRKHSRHEE---NEYHWERKRNSDNPVKVYNSMGSTAEVDESKLTSSEVEPD 837
Query: 262 EVEKDRLHGEEEEEREVSVVNEEKELKHGKELE 294
++E+D L E+EE +++ + EE + +E
Sbjct: 838 DLEEDTLQLPEQEEEDLNRIKEESRRRREAIME 936
Score = 30.4 bits (67), Expect = 1.3
Identities = 43/206 (20%), Positives = 81/206 (38%), Gaps = 25/206 (12%)
Frame = +1
Query: 170 RDEIDEDGEEEREADGLHVDEENERE----------VDDGNEETELKYDKGYEMVRILVN 219
R IDE +R DEE R VDD E + +++ +
Sbjct: 94 RSIIDEYAHSKRRRSDYDHDEERVRVRGREHGHGSVVDDRREYSTRYHNREARDRDRSRD 273
Query: 220 HNVLKEQYMDKYKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEE----- 274
+V ++ + +K KQE+ K + E+ E ER + EKDR EE+
Sbjct: 274 RDVDRDLHREK-KQEETSRGKEIEWEHKRGKERERSRERYRRDAEKDRSREREEDGDRRQ 450
Query: 275 ----EREVSVVNE-----EKELKHGKELEMNISRELE-SGIRDGLDGDEEEEREGDEEKE 324
+R V E EKE ++ + R+ + RD + + ++R G ++++
Sbjct: 451 EKERDRSWDTVFERDRRREKERDRSRDRTIGGKRDRDPENERDDKNRERRDDRYGHKDRD 630
Query: 325 VKHVKEKKKIEKLEKENEEKKVKRGK 350
+ K++ + +N ++ K +
Sbjct: 631 TANGKDRHLRHEDGNDNGDRYRKHSR 708
>TC207029 weakly similar to UP|Q7R8E7 (Q7R8E7) Ran-binding protein, partial
(19%)
Length = 712
Score = 49.7 bits (117), Expect = 2e-06
Identities = 32/105 (30%), Positives = 63/105 (59%), Gaps = 3/105 (2%)
Frame = +1
Query: 250 DRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGL 309
D ++E+ + + GE +++ ++++++E +++++ + GKE ++E +RD +
Sbjct: 16 DTLKEKGKNDEGEDDEENKKNKKKDKKEKEKDHKKEKKEDGKEEGEKEKSKVEVSVRD-I 192
Query: 310 DGDE---EEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKRGKD 351
D +E E E+E E+ + K VKEKKK E +K+ +EKK GKD
Sbjct: 193 DIEETTKEGEKEDKEKDDGKEVKEKKKKE--DKDKKEKKKVTGKD 321
Score = 40.8 bits (94), Expect = 0.001
Identities = 24/102 (23%), Positives = 53/102 (51%)
Frame = +1
Query: 247 DGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIR 306
+G D E ++ ++ + EK++ H +E++E +EK +++I + G +
Sbjct: 46 EGEDDEENKKNKKKDKKEKEKDHKKEKKEDGKEEGEKEKSKVEVSVRDIDIEETTKEGEK 225
Query: 307 DGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKR 348
+ + D+ +E + ++KE K KEKKK+ +K + +K+
Sbjct: 226 EDKEKDDGKEVKEKKKKEDKDKKEKKKVTGKDKTKDLSALKQ 351
Score = 39.3 bits (90), Expect = 0.003
Identities = 29/101 (28%), Positives = 49/101 (47%)
Frame = +1
Query: 27 ESGLRDRRDGDGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMVRKCRRAGL 86
E G D + D E + K K +EKE DH K+ + K + ++ K E VR
Sbjct: 28 EKGKNDEGEDDEENKKNKKKDKKEKEKDHKKEKKEDGKEEGEKEKSKVEVSVR------- 186
Query: 87 DGDGEKEREAGEDLESGRRDGLDGYGEEEREIGEVDEEKEV 127
D D E+ + GE + + DG + ++++E + E+K+V
Sbjct: 187 DIDIEETTKEGEKEDKEKDDGKEVKEKKKKEDKDKKEKKKV 309
Score = 32.0 bits (71), Expect = 0.43
Identities = 39/168 (23%), Positives = 76/168 (45%), Gaps = 11/168 (6%)
Frame = +1
Query: 179 EEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYKQEKRYN 238
++ E D L +N D+G ++ E K +K + +H K++ + K+E
Sbjct: 1 KDEETDTLKEKGKN----DEGEDDEENKKNKKKDKKEKEKDH---KKEKKEDGKEEGEKE 159
Query: 239 SKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKH--GKELEMN 296
+ V +D E +E E + EKD G+E +E++ ++KE K GK+ +
Sbjct: 160 KSKVEVSVRDIDIEETTKEGEKEDKEKD--DGKEVKEKKKKEDKDKKEKKKVTGKDKTKD 333
Query: 297 IS------RELESGIRDGLDGDEEEER---EGDEEKEVKHVKEKKKIE 335
+S ++ I+ L+ + ER E + E V + K+K +++
Sbjct: 334 LSALKQKLEKINGKIQPLLEKKADIERQIKEAEAEGHVVNGKDKDEVQ 477
Score = 28.5 bits (62), Expect = 4.8
Identities = 31/172 (18%), Positives = 73/172 (42%)
Frame = +1
Query: 82 RRAGLDGDGEKEREAGEDLESGRRDGLDGYGEEEREIGEVDEEKEVKLGKKFGMAWIQVN 141
+ G + +GE + E ++ + +++ + +E++E G+ + EKE K ++ ++
Sbjct: 25 KEKGKNDEGEDDEENKKNKKKDKKEKEKDHKKEKKEDGKEEGEKE---KSKVEVSVRDID 195
Query: 142 HYELREHGERRKCGLLEFNFSIDLDSGIRDEIDEDGEEEREADGLHVDEENEREVDDGNE 201
E + GE+ D+ +DG+E +E ++ E++ G +
Sbjct: 196 IEETTKEGEK------------------EDKEKDDGKEVKEKKKKEDKDKKEKKKVTGKD 321
Query: 202 ETELKYDKGYEMVRILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDGLDRVE 253
+T+ ++ +I L E+ D +Q K ++ +V D V+
Sbjct: 322 KTKDLSALKQKLEKINGKIQPLLEKKADIERQIKEAEAEGHVVNGKDKDEVQ 477
>TC206106 weakly similar to UP|Q7RK82 (Q7RK82) Maebl (Fragment), partial (7%)
Length = 1040
Score = 49.7 bits (117), Expect = 2e-06
Identities = 50/208 (24%), Positives = 101/208 (48%), Gaps = 23/208 (11%)
Frame = +2
Query: 167 SGIRDEIDE---DGEEEREADGLHVDEE-NEREVDDGNEETELKYDKGY-EMVRILVNHN 221
SG+ D+I + + E E EA+ ++E E E E ++ + E ++ +
Sbjct: 392 SGVEDKISQSEAEAEAEVEANAKSIEEVVAEAEKAQSGAEDKISQSVSFKEETNVVGDLP 571
Query: 222 VLKEQYMDKYKQ--EKRYNSKNLIV----------EYDGLDRVEEE--EEREVGEVEKDR 267
+ + +D+ K+ ++ N+ L E L++ EEE EE++ EV +++
Sbjct: 572 EAQRKALDELKRLVQEALNNHELTAPKPEPEKKKTELKALEKKEEEVSEEKKEVEVTEEK 751
Query: 268 LHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDEEEEREGDE----EK 323
E EE++ + V EEK KE+E+ ++ G + + + EE++ E +K
Sbjct: 752 KEAEVTEEKKEAEVTEEK-----KEVEVAEEKKEVEGTEEKKEVEVAEEKKETEVIKEKK 916
Query: 324 EVKHVKEKKKIEKLEKENEEKKVKRGKD 351
EV+ +EKK++E E++ E + ++ K+
Sbjct: 917 EVEVTEEKKEVEVTEEKKEAEVIEEKKE 1000
Score = 47.0 bits (110), Expect = 1e-05
Identities = 46/183 (25%), Positives = 89/183 (48%), Gaps = 6/183 (3%)
Frame = +2
Query: 175 EDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLK--EQYMDKYK 232
+ G E++ + + EE V D E D+ +V+ +N++ L + +K K
Sbjct: 497 QSGAEDKISQSVSFKEETN-VVGDLPEAQRKALDELKRLVQEALNNHELTAPKPEPEKKK 673
Query: 233 QEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGE-EEEEREVSVVNEEKELK--- 288
E + K + VE EE++ EV +++ E EE++EV V E+KE++
Sbjct: 674 TELKALEKKEEEVSEEKKEVEVTEEKKEAEVTEEKKEAEVTEEKKEVEVAEEKKEVEGTE 853
Query: 289 HGKELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKR 348
KE+E+ ++ I++ ++E +E+KEV+ +EKK+ E +E++ E +
Sbjct: 854 EKKEVEVAEEKKETEVIKE-----KKEVEVTEEKKEVEVTEEKKEAEVIEEKKEVDATEE 1018
Query: 349 GKD 351
K+
Sbjct: 1019KKE 1027
>TC224025 weakly similar to UP|Q6HXC4 (Q6HXC4) Conserved domain protein,
partial (15%)
Length = 614
Score = 49.3 bits (116), Expect = 3e-06
Identities = 58/193 (30%), Positives = 91/193 (47%), Gaps = 9/193 (4%)
Frame = -1
Query: 156 LLEFNFSIDLDSGIRDEIDEDGEEE-READGLHVDEE-NEREVDDGNEET-ELKYDKGYE 212
+++ N S D + ++ E G+EE ++ +G EE E+E + G EE EL +KG E
Sbjct: 533 IVDLNISADNEDCTGEKEGEKGKEELKDQEGDQGKEELKEQEGEKGKEELRELDAEKGKE 354
Query: 213 MVRILVNHNVLKEQYMDKYKQEKRYNSKNLIVEYDGLDRVEE--EEEREVGEVEKDRLHG 270
+R L E+ ++ K+ K + E DG EE E + E G+ E L G
Sbjct: 353 ELRELD-----AEKAKEELKELDGEKGKEELKELDGEKGKEELKELDGEKGKEELKELDG 189
Query: 271 EEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGD----EEEEREGDEEKEVK 326
E+ +E E E + GKE + + G L+G E +E+EGD+ KE
Sbjct: 188 EKGKEELT-----ELEGQKGKEELTELEGQKGKGELTELEGQKGKVELKEQEGDKGKE-- 30
Query: 327 HVKEKKKIEKLEK 339
K K+++LEK
Sbjct: 29 ----KLKVQELEK 3
>TC210783 weakly similar to UP|O14760 (O14760) Small intestinal mucin MUC3
(Fragment), partial (5%)
Length = 735
Score = 47.0 bits (110), Expect = 1e-05
Identities = 45/180 (25%), Positives = 87/180 (48%), Gaps = 5/180 (2%)
Frame = +3
Query: 177 GEEEREADGLHVDEEN----EREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYK 232
GEEE + D E+ +D+ E+ +L+ D G ++L N+ +
Sbjct: 207 GEEESVNEVSKTDSNGKSFQEKTLDEITEKKDLETDGG----KVLENNGI---------- 344
Query: 233 QEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKE 292
KN++ E +E+++ V +VE+D+ +G E+ +E +E +K KE
Sbjct: 345 ------KKNIVKE------IEDKKAVGVEKVEEDKKNGVVEDAKEDK---KEDGVKEAKE 479
Query: 293 LEMNISREL-ESGIRDGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKRGKD 351
+ + E+ E DG++ +E ++E D +EVK VKE +++++++ E VK K+
Sbjct: 480 DKKEVVEEVKEDNKEDGVEEAKENKKE-DGVEEVKEVKEVDGVKEIKEDKEVDGVKEVKE 656
Score = 31.2 bits (69), Expect = 0.74
Identities = 28/108 (25%), Positives = 51/108 (46%), Gaps = 10/108 (9%)
Frame = +3
Query: 35 DGDGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMVRKCRRAGLDGDGEKER 94
DG + +K N KE++ K + + K++ ++ + E + G+ E ++
Sbjct: 312 DGGKVLENNGIKKNIVKEIEDKKAVGV-EKVEEDKKNGVVEDAKEDKKEDGVKEAKEDKK 488
Query: 95 EAGEDL-ESGRRDGL---------DGYGEEEREIGEVDEEKEVKLGKK 132
E E++ E + DG+ DG EE +E+ EVD KE+K K+
Sbjct: 489 EVVEEVKEDNKEDGVEEAKENKKEDGV-EEVKEVKEVDGVKEIKEDKE 629
>CO985236
Length = 466
Score = 45.1 bits (105), Expect = 5e-05
Identities = 48/174 (27%), Positives = 85/174 (48%), Gaps = 3/174 (1%)
Frame = -1
Query: 175 EDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYKQE 234
E+ E+++ DG V+E NE +DG E+K DK + V + +KE D
Sbjct: 427 EEATEDKKEDG--VEEVNEDRKEDG--VGEVKEDKEVDGV------SCVKEVEEDG---- 290
Query: 235 KRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELE 294
E DG+ +VE+++ E+ E+E+D+ +++ E ++E+ E+K E E
Sbjct: 289 ----------EVDGVKKVEDKKGDELKEIEEDK---KDDHISENDKMDEDTEIKETMEGE 149
Query: 295 -MNISRELESGIRDGL--DGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKK 345
N E E + D + +G +E+ E E ++V+ V E+ EK+ +E K
Sbjct: 148 PENGKEESEKPVVDAMEVEGGIKEKEESKENEKVEVVMEED-----EKDEDEDK 2
Score = 40.8 bits (94), Expect = 0.001
Identities = 30/101 (29%), Positives = 55/101 (53%), Gaps = 3/101 (2%)
Frame = -1
Query: 250 DRVEEEEEREVGEVEKDRLH---GEEEEEREVSVVNEEKELKHGKELEMNISRELESGIR 306
+ E+++E V EV +DR GE +E++EV V+ KE++ E G
Sbjct: 424 EATEDKKEDGVEEVNEDRKEDGVGEVKEDKEVDGVSCVKEVE-------------EDGEV 284
Query: 307 DGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVK 347
DG+ + E+++GDE KE++ K+ I + +K +E+ ++K
Sbjct: 283 DGVK--KVEDKKGDELKEIEEDKKDDHISENDKMDEDTEIK 167
>AW396581
Length = 401
Score = 45.1 bits (105), Expect = 5e-05
Identities = 32/114 (28%), Positives = 61/114 (53%), Gaps = 5/114 (4%)
Frame = -1
Query: 243 IVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELE 302
+ E+DG D E++E+++ + +KD + + E+ ++EK+ K K + E
Sbjct: 368 VKEHDGEDAEEKKEKKKKEKKDKD---DKTDVEKVKGKEDKEKKKKEKKVKDDKTDAEKV 198
Query: 303 SGIRDGLDGDEEEEREGDEEKEVKHVKEKKKIEKL-----EKENEEKKVKRGKD 351
G D DG++ EE+E ++K+ K +K +EK+ + E+E KK K+ K+
Sbjct: 197 KGKED--DGEDNEEKEEKKKKKKKEKDDKIDVEKVKGKEDDGEDEGKKEKKKKE 42
Score = 37.7 bits (86), Expect = 0.008
Identities = 30/103 (29%), Positives = 50/103 (48%), Gaps = 5/103 (4%)
Frame = -1
Query: 35 DGDGEEDREKVKVNEEKEVDHGKKLEMWNKMDYKERKKIREKMVRKCRR-----AGLDGD 89
DG+ E++++ K E+K+ D +E + KE+KK +EK V+ + G + D
Sbjct: 356 DGEDAEEKKEKKKKEKKDKDDKTDVEKVKGKEDKEKKK-KEKKVKDDKTDAEKVKGKEDD 180
Query: 90 GEKEREAGEDLESGRRDGLDGYGEEEREIGEVDEEKEVKLGKK 132
GE E E + +++ D E+ + E D E E K KK
Sbjct: 179 GEDNEEKEEKKKKKKKEKDDKIDVEKVKGKEDDGEDEGKKEKK 51
Score = 28.1 bits (61), Expect = 6.2
Identities = 19/60 (31%), Positives = 32/60 (52%), Gaps = 5/60 (8%)
Frame = -1
Query: 4 EKRMKNENTRC-KMKNEKPFSSDLESG----LRDRRDGDGEEDREKVKVNEEKEVDHGKK 58
EK++K++ T K+K ++ D E + +++ D + D EKVK E+ D GKK
Sbjct: 239 EKKVKDDKTDAEKVKGKEDDGEDNEEKEEKKKKKKKEKDDKIDVEKVKGKEDDGEDEGKK 60
>CF921229
Length = 680
Score = 44.7 bits (104), Expect = 6e-05
Identities = 31/101 (30%), Positives = 60/101 (58%), Gaps = 8/101 (7%)
Frame = -3
Query: 252 VEEEEEREVGEVEKDRLHGE-EEEEREVSVVNEEKE----LKHGKELEMNISR---ELES 303
+E EE++ EV++++ GE EE++EV V E+KE ++ KE+E+ + E+
Sbjct: 615 IEVTEEKKEAEVKEEKKEGEVTEEKKEVEVTEEKKEAEVIVEEKKEVEVTEEKKEVEVTE 436
Query: 304 GIRDGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEK 344
G ++ +E++E E EEK+ V+ +++ ++ E + EEK
Sbjct: 435 GKKEVEVIEEKKETEVTEEKKEVEVEVREEKKESEVKEEEK 313
Score = 41.6 bits (96), Expect = 5e-04
Identities = 30/100 (30%), Positives = 55/100 (55%)
Frame = -3
Query: 252 VEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDG 311
+EE++E EV E +K+ EE++E EV +E E+ K+ E+ ++ E
Sbjct: 660 IEEKKEVEVTEEKKEIEVTEEKKEAEVKEEKKEGEVTEEKK-EVEVTEE----------- 517
Query: 312 DEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVKRGKD 351
+E E +E+KEV+ +EKK++E E + E + ++ K+
Sbjct: 516 KKEAEVIVEEKKEVEVTEEKKEVEVTEGKKEVEVIEEKKE 397
Score = 39.3 bits (90), Expect = 0.003
Identities = 32/99 (32%), Positives = 56/99 (56%), Gaps = 8/99 (8%)
Frame = -3
Query: 253 EEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELK---HGKELEMNISRELESGIRDGL 309
EE++E EV E +K+ EE++E EV +V E+KE++ KE+E+ ++ I +
Sbjct: 576 EEKKEGEVTEEKKEVEVTEEKKEAEV-IVEEKKEVEVTEEKKEVEVTEGKKEVEVIEEKK 400
Query: 310 DGDEEEER-----EGDEEKEVKHVKEKKKIEKLEKENEE 343
+ + EE+ E EEK+ VKE++K +++ E E
Sbjct: 399 ETEVTEEKKEVEVEVREEKKESEVKEEEKGQEVVPEEVE 283
Score = 39.3 bits (90), Expect = 0.003
Identities = 60/254 (23%), Positives = 112/254 (43%), Gaps = 24/254 (9%)
Frame = -3
Query: 113 EEEREIGEVDEEKEVKLGKKFGMAWIQVNHYELREHGERRKCGLLEFNFSIDLDSGIRDE 172
EE++E+ +E+KE+++ ++ A ++ E E+++
Sbjct: 657 EEKKEVEVTEEKKEIEVTEEKKEAEVKEEKKEGEVTEEKKEV------------------ 532
Query: 173 IDEDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYK 232
E EE++EA+ + V+E+ E EV + +E E+ K KE + + K
Sbjct: 531 --EVTEEKKEAEVI-VEEKKEVEVTEEKKEVEVTEGK--------------KEVEVIEEK 403
Query: 233 QEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDR-LHGEEEEEREVSVVNEEKE----L 287
+E + VE ++ EE++E EV E EK + + EE E + ++ +E+ L
Sbjct: 402 KETEVTEEKKEVE---VEVREEKKESEVKEEEKGQEVVPEEVEIWGIPLLGDERSDVILL 232
Query: 288 KHGKELEMNISRELESGIRD--------GLDGDEEEEREGD-----------EEKEVKHV 328
K + + + L IR+ G++G EE+ D E+KEV+
Sbjct: 231 KFLRARDFKVKEALNM-IRNTVRWRKEFGIEGLVEEDLGSD*EKKEAEVIVEEKKEVEVT 55
Query: 329 KEKKKIEKLEKENE 342
+EKK++E E + E
Sbjct: 54 EEKKEVEVTEGKKE 13
>BU763591
Length = 389
Score = 43.9 bits (102), Expect = 1e-04
Identities = 33/100 (33%), Positives = 57/100 (57%), Gaps = 1/100 (1%)
Frame = -1
Query: 252 VEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDG 311
+E EE++ EV++++ GE EE++ V EEK KE E+ + + E + + +
Sbjct: 287 IEVTEEKKEAEVKEEKKEGEVTEEKKEVEVTEEK-----KEAEVIVEEKKEVEVTE--EK 129
Query: 312 DEEEEREGDEEKEVKHVKEKKKIEKLEKENE-EKKVKRGK 350
E E EG +KEV+ ++EKK+ E E++ E E +V+ K
Sbjct: 128 KEVEVTEG--KKEVEVIEEKKETEVTEEKKEVEVEVREEK 15
Score = 42.7 bits (99), Expect = 2e-04
Identities = 31/99 (31%), Positives = 55/99 (55%), Gaps = 3/99 (3%)
Frame = -1
Query: 255 EEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDEE 314
EE++E+ E+ + EE++EV V E+KE++ +E +E E +E
Sbjct: 383 EEKKEIEVTEEKKKAEVIEEKKEVEVTEEKKEIEVTEE-----KKEAEV---------KE 246
Query: 315 EEREGD---EEKEVKHVKEKKKIEKLEKENEEKKVKRGK 350
E++EG+ E+KEV+ +EKK+ E + +E +E +V K
Sbjct: 245 EKKEGEVTEEKKEVEVTEEKKEAEVIVEEKKEVEVTEEK 129
>TC206440
Length = 855
Score = 43.1 bits (100), Expect = 2e-04
Identities = 49/198 (24%), Positives = 87/198 (43%), Gaps = 12/198 (6%)
Frame = +3
Query: 166 DSGIRDEID----EDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYE-MVRILVNH 220
+S I D I+ E + ++ + +DEE + E+ EE E+ E V +
Sbjct: 3 ESSITDVIEAVLNEAADIKKTPEQASIDEEAKTEL---KEEREISVPAAVEEKVAVAAID 173
Query: 221 NVLKEQ-----YMDKYKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEE 275
+V K++ + QE+ + SK RVEE+ E + E + +
Sbjct: 174 DVDKKEPEAPNVSPESSQEEEFESK----------RVEEQNEAKAATTETVE---SVKVD 314
Query: 276 REVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDEEEE--REGDEEKEVKHVKEKKK 333
+ S+V+++KE K ++E ISR + +R+ L EEE G ++ E + E +K
Sbjct: 315 NDSSIVDDKKEEKTDSKVE-EISRAINEPVRETLASKFEEETVEPGADKVEKEQTVEPEK 491
Query: 334 IEKLEKENEEKKVKRGKD 351
E+++ E K KD
Sbjct: 492 TEEVQATKESDATKTSKD 545
>BG649872
Length = 326
Score = 42.0 bits (97), Expect = 4e-04
Identities = 30/100 (30%), Positives = 49/100 (49%)
Frame = -1
Query: 245 EYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESG 304
E DG+ +VE+++ E+ E+E+D+ E ++ E KE GK+ + E
Sbjct: 293 EVDGVKKVEDKKGDELKEIEEDKKDDHISENDKMDEDTEVKETIEGKQENEKVQAEKPEV 114
Query: 305 IRDGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKENEEK 344
++G +EE E DE KEKK + E E+E+K
Sbjct: 113 DAMEVEGGIQEE-ESDE-------KEKKDVAMEEDEDEDK 18
Score = 29.3 bits (64), Expect = 2.8
Identities = 20/62 (32%), Positives = 33/62 (52%), Gaps = 1/62 (1%)
Frame = -1
Query: 283 EEKELKHGKELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKH-VKEKKKIEKLEKEN 341
E+KE+ K++E EL+ D D E + DE+ EVK ++ K++ EK++ E
Sbjct: 302 EDKEVDGVKKVEDKKGDELKEIEEDKKDDHISENDKMDEDTEVKETIEGKQENEKVQAEK 123
Query: 342 EE 343
E
Sbjct: 122 PE 117
Score = 28.5 bits (62), Expect = 4.8
Identities = 12/33 (36%), Positives = 23/33 (69%)
Frame = -1
Query: 315 EEREGDEEKEVKHVKEKKKIEKLEKENEEKKVK 347
E+++GDE KE++ K+ I + +K +E+ +VK
Sbjct: 269 EDKKGDELKEIEEDKKDDHISENDKMDEDTEVK 171
>BU927182
Length = 447
Score = 41.6 bits (96), Expect = 5e-04
Identities = 38/126 (30%), Positives = 62/126 (49%)
Frame = +3
Query: 169 IRDEIDEDGEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYM 228
++ E ++ E+E E + + V E++ RE + E+ + +K E R V K++
Sbjct: 51 LQKERKKEFEKEHEGERVRVREKDRREREKEVEKERREREKEVEKDRREREKEVEKDRRE 230
Query: 229 DKYKQEKRYNSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELK 288
+ + EK K VE D R E+E+E VEKDR ERE VV + K+++
Sbjct: 231 REKEVEKDRREKEKEVEKD---RREKEKE-----VEKDR------REREKEVVKDRKKIE 368
Query: 289 HGKELE 294
+ KE E
Sbjct: 369 NDKERE 386
Score = 37.7 bits (86), Expect = 0.008
Identities = 31/103 (30%), Positives = 52/103 (50%), Gaps = 1/103 (0%)
Frame = +3
Query: 250 DRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGL 309
+R +E E+ GE + R E E+EV E+E KE+E + RE E +
Sbjct: 60 ERKKEFEKEHEGERVRVREKDRREREKEVEKERRERE----KEVEKD-RREREKEVEKDR 224
Query: 310 DGDEEEEREGDEEKEVKHVKEKKKIEK-LEKENEEKKVKRGKD 351
E+E + EKE + K++++ EK +EK+ E++ + KD
Sbjct: 225 REREKEVEKDRREKEKEVEKDRREKEKEVEKDRREREKEVVKD 353
>TC207028 similar to UP|Q9LW95 (Q9LW95) KED, partial (13%)
Length = 490
Score = 40.8 bits (94), Expect = 0.001
Identities = 38/98 (38%), Positives = 47/98 (47%), Gaps = 21/98 (21%)
Frame = +1
Query: 275 EREVSVVNEEKELKHGKEL--EMNISRELESGIRDGLDGDEEEEREGDE-------EKEV 325
ERE V EEK+ K KE E E +++ DE E+ EG++ EKE
Sbjct: 127 EREKMVSGEEKKEKKEKEKKKEKKDKDEETDTLKEKGKNDEGEDDEGNKKKKKDKKEKEK 306
Query: 326 KHVKEKKKIEKLEKEN------------EEKKVKRGKD 351
H KEKK IE+ EKE+ EEKKV GKD
Sbjct: 307 DHKKEKKDIEEGEKEDSKVEASVRDIDIEEKKV-TGKD 417
Score = 31.2 bits (69), Expect = 0.74
Identities = 30/116 (25%), Positives = 62/116 (52%), Gaps = 1/116 (0%)
Frame = +1
Query: 177 GEEEREADGLHVDEENEREVDDGNEETELKYDKGYEMVRILVNHNVLKEQYMDKYKQEKR 236
GEE++E ++E ++E D +EET+ +KG N K++ DK ++EK
Sbjct: 148 GEEKKEKK----EKEKKKEKKDKDEETDTLKEKGKNDEGEDDEGN--KKKKKDKKEKEKD 309
Query: 237 Y-NSKNLIVEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGK 291
+ K I E + D E R++ ++E+ ++ G +++ +++S + ++ E +GK
Sbjct: 310 HKKEKKDIEEGEKEDSKVEASVRDI-DIEEKKVTG-KDKTKDLSTLKQKLEKINGK 471
Score = 30.4 bits (67), Expect = 1.3
Identities = 19/41 (46%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Frame = +3
Query: 312 DEEEEREGDEEKEVKHVKEKKKIEKLEKENEEKKVK-RGKD 351
D+ E+ E E+KE K KEKKK EK +K+ E +K +GK+
Sbjct: 21 DKGEDGEEKEKKE-KDKKEKKKKEKKDKDEETDTLKGKGKN 140
>TC226049 similar to UP|O24078 (O24078) Protein phosphatase 2C, partial (34%)
Length = 604
Score = 40.4 bits (93), Expect = 0.001
Identities = 30/99 (30%), Positives = 48/99 (48%), Gaps = 1/99 (1%)
Frame = -2
Query: 244 VEYDGLDRVEEEEEREVGEVEKDRLHGEEEEEREVSVVNEEKELKHGKELE-MNISRELE 302
V + +D EEE E VG + + +GEE EE E + E++ +ELE + RE E
Sbjct: 399 VPFGEVDEAEEEAENTVGGLGRRSRNGEEGEEDEEAAAEGEEQDDVVEELEGLRCERESE 220
Query: 303 SGIRDGLDGDEEEEREGDEEKEVKHVKEKKKIEKLEKEN 341
D EEE EG E+K + + ++E +++
Sbjct: 219 R------DSGEEEMMEGFEQKREEEGENTGELETATEQD 121
>TC203614 UP|Q94LX2 (Q94LX2) Beta-conglycinin alpha subunit, complete
Length = 1983
Score = 40.4 bits (93), Expect = 0.001
Identities = 30/108 (27%), Positives = 56/108 (51%), Gaps = 10/108 (9%)
Frame = +3
Query: 254 EEEEREVGEVEKDRLHGEEEEEREVSVVNEE--KELKHGKELEMNISRELESGIRDGLDG 311
E E ++ GE E+D E+E+ R + + +E +H + E R+ E + G G
Sbjct: 288 EREPQQPGEKEED----EDEQPRPIPFPRPQPRQEEEHEQREEQEWPRKEE---KRGEKG 446
Query: 312 DEEEEREGDEEKEVK--------HVKEKKKIEKLEKENEEKKVKRGKD 351
EEE+ + DEE++ + H KE++K E+ E E ++++ + +D
Sbjct: 447 SEEEDEDEDEEQDERQFPFPRPPHQKEERKQEEDEDEEQQRESEESED 590
>TC215868 similar to UP|Q9LJ56 (Q9LJ56) Gb|AAD43149.1, partial (50%)
Length = 1472
Score = 39.3 bits (90), Expect = 0.003
Identities = 23/63 (36%), Positives = 35/63 (55%)
Frame = +1
Query: 271 EEEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHVKE 330
+EEEE+E N+ E + ++ N + S ++ D DEEEE E +EE + KH K+
Sbjct: 277 QEEEEKEEKHQNDHVE-EQEEDAPNNPEQSQPSDSKEVTDEDEEEEDEEEEEDKPKHAKK 453
Query: 331 KKK 333
KK
Sbjct: 454 AKK 462
>TC214290 homologue to UP|Q6UJX5 (Q6UJX5) Molecular chaperone Hsp90-2,
complete
Length = 2566
Score = 39.3 bits (90), Expect = 0.003
Identities = 28/82 (34%), Positives = 42/82 (51%), Gaps = 2/82 (2%)
Frame = +1
Query: 272 EEEEREVSVVNEEKEL--KHGKELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHVK 329
+E++ E K+L KH + + IS +E + DE+EE + DEE +V+ V
Sbjct: 745 KEDQLEYLEERRLKDLIKKHSEFISYPISLWIEKTTEKEISDDEDEEEKKDEEGKVEDVD 924
Query: 330 EKKKIEKLEKENEEKKVKRGKD 351
E EKE EEKK K+ K+
Sbjct: 925 E-------EKEKEEKKKKKIKE 969
Score = 28.1 bits (61), Expect = 6.2
Identities = 25/110 (22%), Positives = 51/110 (45%), Gaps = 8/110 (7%)
Frame = +1
Query: 145 LREHGERRKCGLLEFNFSIDLDSGIRDEI--DEDGEEEREADGL--HVDEENEREVDDGN 200
+++H E + + S+ ++ EI DED EE+++ +G VDEE E+E
Sbjct: 793 IKKHSE-----FISYPISLWIEKTTEKEISDDEDEEEKKDEEGKVEDVDEEKEKEEKKKK 957
Query: 201 EETELKYD----KGYEMVRILVNHNVLKEQYMDKYKQEKRYNSKNLIVEY 246
+ E+ ++ + + + + KE+Y YK ++L V++
Sbjct: 958 KIKEVSHEWSLVNKQKPIWMRKPEEITKEEYSAFYKSLTNDWEEHLAVKH 1107
>BM527600 similar to PIR|T05883|T058 ATP-dependent helicase F6H11.20 -
Arabidopsis thaliana, partial (8%)
Length = 422
Score = 38.9 bits (89), Expect = 0.004
Identities = 24/76 (31%), Positives = 39/76 (50%)
Frame = +1
Query: 272 EEEEREVSVVNEEKELKHGKELEMNISRELESGIRDGLDGDEEEEREGDEEKEVKHVKEK 331
E++ + S +NE K++K K + + + + +EE E D + E K+K
Sbjct: 7 EDKTLQASPMNETKKMKKNKNKKKSDKKRPREIATEERPEEEENENNTDSDGESSKKKKK 186
Query: 332 KKIEKLEKENEEKKVK 347
KK+E E E +EKKVK
Sbjct: 187 KKVEG-ESEVKEKKVK 231
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.306 0.133 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,482,737
Number of Sequences: 63676
Number of extensions: 141068
Number of successful extensions: 2506
Number of sequences better than 10.0: 243
Number of HSP's better than 10.0 without gapping: 1637
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2122
length of query: 351
length of database: 12,639,632
effective HSP length: 98
effective length of query: 253
effective length of database: 6,399,384
effective search space: 1619044152
effective search space used: 1619044152
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0315.14


