

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
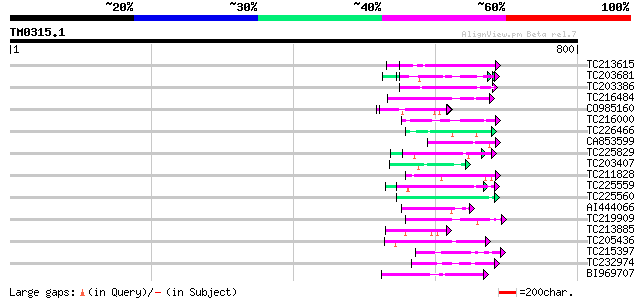
Query= TM0315.1
(800 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC213615 similar to UP|Q9SUL7 (Q9SUL7) SNF1 like protein kinase ... 54 4e-07
TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 50 3e-06
TC203386 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 48 1e-05
TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 47 3e-05
CO985160 47 4e-05
TC216000 similar to UP|O22253 (O22253) Photolyase/blue-light rec... 46 6e-05
TC226466 weakly similar to GB|AAD49970.1|5734705|F24J5 F24J5.4 {... 46 7e-05
CA853599 weakly similar to PIR|F86379|F86 protein F21J9.28 [impo... 46 7e-05
TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like ara... 46 7e-05
TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-pr... 45 1e-04
TC211828 similar to UP|RPIA_ARATH (Q9ZU38) Probable-ribose 5-pho... 45 1e-04
TC225559 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 45 1e-04
TC225560 similar to UP|Q9FUL8 (Q9FUL8) Arabinogalactan protein, ... 45 1e-04
AI444066 similar to GP|9294409|dbj polygalacturonase inhibitor-l... 44 2e-04
TC219909 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-... 44 3e-04
TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partia... 44 3e-04
TC205436 homologue to UP|PRS6_ARATH (Q9SEI4) 26S protease regula... 44 4e-04
TC215397 weakly similar to UP|Q8S9J6 (Q8S9J6) AT5g10770/T30N20_4... 44 4e-04
TC232974 weakly similar to GB|AAK54457.1|20384750|AY032998 DNA r... 43 5e-04
BI969707 43 6e-04
>TC213615 similar to UP|Q9SUL7 (Q9SUL7) SNF1 like protein kinase
(CBL-interacting protein kinase 4), partial (45%)
Length = 674
Score = 53.5 bits (127), Expect = 4e-07
Identities = 37/143 (25%), Positives = 66/143 (45%)
Frame = +3
Query: 550 AGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAA 609
A T+ P + +T ++P S +++T P A ASS S+ A AA + S + +
Sbjct: 258 ASTTTPTSSKSTRSSPPRPR----STSSSTSPVA--ASSSPSSPAAAASRSPSPAATSRS 419
Query: 610 SAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCP 669
S+ ++A+ A +S T + TP + R SPPS + + ++ S P + P
Sbjct: 420 SSPLSASATAMASPTETSSHRTSSSTPPATSRSPTSASPPSRNTSTTVSSTPPAARRPSP 599
Query: 670 GAATTSEAAQIEQAPAPEVGSSS 692
+++ +A P P +SS
Sbjct: 600 RQRSSAASATTAXKPTPGPAASS 668
Score = 45.1 bits (105), Expect = 1e-04
Identities = 46/162 (28%), Positives = 69/162 (42%), Gaps = 1/162 (0%)
Frame = +3
Query: 532 AESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPAS 591
A ST P + T++S S + AA+ + A A+ + + +SSP S
Sbjct: 255 AASTTTPTSSKSTRSSPPRPRSTSSSTSPVAASSSPSSPAAAASRSPSPAATSRSSSPLS 434
Query: 592 AGATA-AVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPS 650
A ATA A E+S T++S A+S +P T SPP +N + S
Sbjct: 435 ASATAMASPTETSSHRTSSST------PPATSRSP---------TSASPPSRNTSTTVSS 569
Query: 651 THDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSS 692
T A PSP ++S +ATT+ + P P SS+
Sbjct: 570 TPPAARRPSP---RQRSSAASATTA----XKPTPGPAASSST 674
Score = 31.6 bits (70), Expect = 1.4
Identities = 44/208 (21%), Positives = 74/208 (35%), Gaps = 7/208 (3%)
Frame = +3
Query: 492 EDMNFTSAELLKARKRRMARFSPKFNTEGDAKKRGGAENQAESTKAPKRRRLTKASSDAG 551
E ++F + K R+RR R + + + A +T RR +++
Sbjct: 6 EAIHFHFPQQWKHRRRRHRR---RHHGPPSWPSTSSRVSSAAATSLRSTRRAPSSTAPR* 176
Query: 552 TSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASA 611
P P P S A T P SS ++ + + SS + A++
Sbjct: 177 P*RPSTSPRRWTPPWSPASCVRSTPCAAS-TTTPTSSKSTRSSPPRPRSTSSSTSPVAAS 353
Query: 612 GVNATKAAASSDTPIGDEEKENETPKSPPRQN-APPSPPSTHDAGSMP------SPPHQG 664
++ AAA+S +P + +P S A P+ S+H S SP
Sbjct: 354 SSPSSPAAAASRSPSPAATSRSSSPLSASATAMASPTETSSHRTSSSTPPATSRSPTSAS 533
Query: 665 EKSCPGAATTSEAAQIEQAPAPEVGSSS 692
S + T S + P+P SS+
Sbjct: 534 PPSRNTSTTVSSTPPAARRPSPRQRSSA 617
>TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 913
Score = 50.4 bits (119), Expect = 3e-06
Identities = 43/141 (30%), Positives = 61/141 (42%)
Frame = +3
Query: 551 GTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAAS 610
G +P A PTT+ AA T P A PA++P+ + A VA+ +S A+S
Sbjct: 69 GGQSPAASPTTSPP-----------AATTTPFASPATAPSKPKSPAPVASPTSSSPPASS 215
Query: 611 AGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPG 670
NA A + +P T SPP + A P+P +T A + P+ + P
Sbjct: 216 P--NAATATPPASSP---------TVASPPSKAAAPAPVATPPAATPPA-------ATPP 341
Query: 671 AATTSEAAQIEQAPAPEVGSS 691
AAT + PAP SS
Sbjct: 342 AATPPAVTPVSSPPAPVPVSS 404
Score = 45.1 bits (105), Expect = 1e-04
Identities = 41/152 (26%), Positives = 62/152 (39%), Gaps = 16/152 (10%)
Frame = +3
Query: 546 ASSDAGTSNPGAQPTTAAAPKGKNVAEASVA-------------AATEPTAVPASSPASA 592
+S +A T+ P A T A+P K A A VA AAT P P SSP +
Sbjct: 210 SSPNAATATPPASSPTVASPPSKAAAPAPVATPPAATPPAATPPAATPPAVTPVSSPPAP 389
Query: 593 GATAAVAAESSIGATAASAGVN-ATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPST 651
++ A + + A A A A S++ P + + +T KS + AP PS
Sbjct: 390 VPVSSPPAPVPVSSPPALAPTTPAPVVAPSAEVPAPAPKSKKKTKKS-KKHTAPAPSPSL 566
Query: 652 HDAGSMP--SPPHQGEKSCPGAATTSEAAQIE 681
+ P +P + PG A + + + E
Sbjct: 567 LGPPAPPVGAPGSSQDSMSPGPAVSEDESGAE 662
Score = 43.5 bits (101), Expect = 4e-04
Identities = 45/163 (27%), Positives = 61/163 (36%)
Frame = +3
Query: 526 GGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVP 585
GG A T +P T +S A + P A+P + +S AAT P
Sbjct: 69 GGQSPAASPTTSPPAATTTPFASPATAPSKPKSPAPVASPTSSSPPASSPNAAT--ATPP 242
Query: 586 ASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAP 645
ASSP A + AA + + A+ AT AA+ PP
Sbjct: 243 ASSPTVASPPSKAAAPAPVATPPAATPPAATPPAAT-----------------PPAVTPV 371
Query: 646 PSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEV 688
SPP+ S P+P S P A T+ A + AP+ EV
Sbjct: 372 SSPPAPVPVSSPPAP--VPVSSPPALAPTTPAPVV--APSAEV 488
>TC203386 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 649
Score = 48.1 bits (113), Expect = 1e-05
Identities = 39/138 (28%), Positives = 56/138 (40%)
Frame = +1
Query: 551 GTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAAS 610
G +P A PTT++A ++ + P A P SSP ++ AA A T AS
Sbjct: 64 GGQSPAAAPTTSSASPATAPSKPKPKSPA-PVASPTSSPPASSPNAATATPPVSSPTVAS 240
Query: 611 AGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPG 670
A A ++ P+ +PP SPP+ S P+P S P
Sbjct: 241 PPSKAAAPAPATTPPVATPPAATPPAATPPAVTPVSSPPAPVPVSSPPAP--VPVSSPPA 414
Query: 671 AATTSEAAQIEQAPAPEV 688
A T+ A + AP+ EV
Sbjct: 415 LAPTTPAPVV--APSAEV 462
Score = 41.6 bits (96), Expect = 0.001
Identities = 38/151 (25%), Positives = 58/151 (38%), Gaps = 15/151 (9%)
Frame = +1
Query: 546 ASSDAGTSNPGAQPTTAAAPKGKNVAEASVA-------------AATEPTAVPASSPASA 592
+S +A T+ P T A+P K A A AAT P P SSP +
Sbjct: 184 SSPNAATATPPVSSPTVASPPSKAAAPAPATTPPVATPPAATPPAATPPAVTPVSSPPAP 363
Query: 593 GATAAVAAESSIGATAASAGVN-ATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPST 651
++ A + + A A A A S++ P + + +T KS PSP
Sbjct: 364 VPVSSPPAPVPVSSPPALAPTTPAPVVAPSAEVPAPAPKSKKKTKKSKKHTAPAPSPALL 543
Query: 652 -HDAGSMPSPPHQGEKSCPGAATTSEAAQIE 681
A + +P + S PG A + + + E
Sbjct: 544 GPPAPPVGAPGSSQDASSPGPAVSEDESGAE 636
>TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (58%)
Length = 1213
Score = 47.0 bits (110), Expect = 3e-05
Identities = 39/152 (25%), Positives = 62/152 (40%), Gaps = 1/152 (0%)
Frame = +1
Query: 534 STKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAG 593
S+ P R T +S T + TT+ +P + A + + P+ P S
Sbjct: 85 SSSPPSPTRTTSPASSQSTPSSPPSTTTSPSPTSPPKSTAKPPSPSAPSTTPPCRTFSRS 264
Query: 594 ATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHD 653
+ + +S +T++S +A +++ S T P SPP+ PP+PP
Sbjct: 265 TPPSTPSRTSSPSTSSST-TSAPRSSTRSPT----------APPSPPQCTKPPAPPPDPP 411
Query: 654 AGSMPSPPHQGEKS-CPGAATTSEAAQIEQAP 684
A S SP EKS P TT+ + Q P
Sbjct: 412 ASS-TSPTSTAEKSHSPPKTTTAHSPQRSSNP 504
>CO985160
Length = 777
Score = 46.6 bits (109), Expect = 4e-05
Identities = 42/117 (35%), Positives = 57/117 (47%), Gaps = 10/117 (8%)
Frame = -1
Query: 518 TEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGT------SNPGAQPTTAAAPK-GKNV 570
T DAK A + A +T A T A+S A T S A T++AP
Sbjct: 735 TSADAKPT--AASSAVATTAAATSHATTATSSAPTTAATTSSAAAAAAATSSAPTTAATT 562
Query: 571 AEASVAAATEPTAVPASSPASAGATAAV---AAESSIGATAASAGVNATKAAASSDT 624
+ A+ AAAT +A A++ S+ ATAA AA + ATAA++ AT AA S+ T
Sbjct: 561 SSAAAAAATTSSAAAAAATTSSAATAAATGPAAAACAAATAAASANPATAAATSTST 391
Score = 44.7 bits (104), Expect = 2e-04
Identities = 37/109 (33%), Positives = 54/109 (48%), Gaps = 7/109 (6%)
Frame = -1
Query: 522 AKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEP 581
A A A ++ AP T +++ A A T++AA + A+ AAAT P
Sbjct: 630 ATTSSAAAAAAATSSAPTTAATTSSAAAA------AATTSSAAAAAATTSSAATAAATGP 469
Query: 582 TAVP-ASSPASAGATAAVAAESSIG------ATAASAGVNATKAAASSD 623
A A++ A+A A A AA +S ATAA++ V AT AAA++D
Sbjct: 468 AAAACAAATAAASANPATAAATSTSTTPTTTATAAASAVTATAAAAATD 322
Score = 35.8 bits (81), Expect = 0.076
Identities = 24/49 (48%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Frame = +2
Query: 573 ASVAAATEPTAVPASSPASAG-ATAAVAAESSIGATAASAGVNATKAAA 620
A+VA E AV A+ A+AG AAVAAE + A AA+ V A AAA
Sbjct: 410 AAVAGFAEAAAVAAAQAAAAGPVAAAVAAEEVVAAAAAAEEVVAAAAAA 556
Score = 35.0 bits (79), Expect = 0.13
Identities = 33/121 (27%), Positives = 53/121 (43%), Gaps = 1/121 (0%)
Frame = -1
Query: 573 ASVAAATEPTAVPASSPASAGATA-AVAAESSIGATAASAGVNATKAAASSDTPIGDEEK 631
A+ +A +PTA ++ +A AT+ A A SS TAA+ A AAA+S P
Sbjct: 741 ATTSADAKPTAASSAVATTAAATSHATTATSSAPTTAATTSSAAAAAAATSSAPTTAATT 562
Query: 632 ENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSS 691
+ + +A + +T A + + G + AA T+ AA A A +S
Sbjct: 561 SSAAAAAATTSSAAAAAATTSSAATAAA---TGPAAAACAAATA-AASANPATAAATSTS 394
Query: 692 S 692
+
Sbjct: 393 T 391
Score = 29.3 bits (64), Expect = 7.1
Identities = 22/67 (32%), Positives = 30/67 (43%)
Frame = +2
Query: 545 KASSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSI 604
+A++ A A P AA AE VAAA V A++ A+ AAV +
Sbjct: 431 EAAAVAAAQAAAAGPVAAAV-----AAEEVVAAAAAAEEVVAAAAAAEEVVAAVVGAEEV 595
Query: 605 GATAASA 611
A AA+A
Sbjct: 596 AAAAAAA 616
>TC216000 similar to UP|O22253 (O22253) Photolyase/blue-light receptor
(Photolyase/blue light photoreceptor PHR2), partial
(84%)
Length = 1641
Score = 46.2 bits (108), Expect = 6e-05
Identities = 41/140 (29%), Positives = 59/140 (41%)
Frame = +3
Query: 553 SNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAG 612
SNP + PT P A AT PT P+++P S+G+ T+AS+
Sbjct: 165 SNPPSPPT----PSTPLSPSLPTAPATLPTPPPSAAPPSSGSAT----------TSASST 302
Query: 613 VNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAA 672
++A+ + TP +P SP + PP+ S H A + P+P S P +
Sbjct: 303 MSASPPPTT--TP---------SPSSPSTASTPPTTASRHPASTRPAPTAPPSSSTP--S 443
Query: 673 TTSEAAQIEQAPAPEVGSSS 692
TS AA AP S S
Sbjct: 444 PTSAAASRRAAPISSCASGS 503
Score = 38.5 bits (88), Expect = 0.012
Identities = 18/56 (32%), Positives = 27/56 (48%)
Frame = +3
Query: 631 KENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAP 686
K P SPP + P SP ++P+PP G+ATTS ++ + +P P
Sbjct: 156 KPPSNPPSPPTPSTPLSPSLPTAPATLPTPPPSAAPPSSGSATTSASSTMSASPPP 323
>TC226466 weakly similar to GB|AAD49970.1|5734705|F24J5 F24J5.4 {Arabidopsis
thaliana;} , partial (22%)
Length = 978
Score = 45.8 bits (107), Expect = 7e-05
Identities = 39/138 (28%), Positives = 55/138 (39%), Gaps = 10/138 (7%)
Frame = +1
Query: 559 PTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKA 618
P+ A AP ASV A + PT + A+SP+S G AV +S T+ ++ A
Sbjct: 46 PSNAQAP-------ASVPANSPPTTLAATSPSS-GTPPAVTQAASPPTTSNPPTISQPPA 201
Query: 619 AASSD----TPIGDEEKENETPKSPPRQNAPPSPPSTHDAGS------MPSPPHQGEKSC 668
+ TP + +PK+P Q PP PP + +P PP
Sbjct: 202 IVAPKPAPVTPPAPKVAPASSPKAPTPQTPPPQPPKVSPVAAPTLPPPLPPPPVATPPPL 381
Query: 669 PGAATTSEAAQIEQAPAP 686
P A+ APAP
Sbjct: 382 PPPKIAPTPAKTSPAPAP 435
>CA853599 weakly similar to PIR|F86379|F86 protein F21J9.28 [imported] -
Arabidopsis thaliana, partial (58%)
Length = 389
Score = 45.8 bits (107), Expect = 7e-05
Identities = 37/111 (33%), Positives = 45/111 (40%), Gaps = 8/111 (7%)
Frame = -1
Query: 590 ASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPP 649
A GA A S T SA + AT AAS+ + TP R + PSP
Sbjct: 389 AGDGAGAGQRRRRSWSWTT*SARLTATAMAASASRSSSSSTQRASTPTRSWRTSRTPSPS 210
Query: 650 STHDAGSMPSPPHQGEKSCPGAATTS--------EAAQIEQAPAPEVGSSS 692
ST A + PSPP SC +A T+ AA + A AP SS
Sbjct: 209 STSTA-TAPSPPRSSTPSCAASARTAPSPSAAG*SAASMATATAPSTSRSS 60
>TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (57%)
Length = 1652
Score = 45.8 bits (107), Expect = 7e-05
Identities = 34/138 (24%), Positives = 51/138 (36%), Gaps = 3/138 (2%)
Frame = +2
Query: 538 PKRRRLTKASSDAGTSNPGAQPTTAAAPKGKN---VAEASVAAATEPTAVPASSPASAGA 594
P T S + TS+P +T +APK A S + P A P + PA++ +
Sbjct: 215 PPSSTSTSPSPPSKTSSPSTSSSTTSAPKSSTRSTTAPPSSPPCSRPPAPPPAPPATSTS 394
Query: 595 TAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDA 654
+ A S+ +A + + S P ++ P +PP PP PP
Sbjct: 395 PTSRPARSASPPKTTTAPSTPSTSNPSRKCPTTSPSSKSAPPLAPPTPRHPPPPP----- 559
Query: 655 GSMPSPPHQGEKSCPGAA 672
PP CP A
Sbjct: 560 -----PPST*SPLCPNKA 598
Score = 42.4 bits (98), Expect = 8e-04
Identities = 38/148 (25%), Positives = 65/148 (43%), Gaps = 16/148 (10%)
Frame = +2
Query: 555 PGAQP--TTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAG 612
P + P TT+ +P + A+ + + P+ +P P+S + + +++S +T++S
Sbjct: 110 PASPPSTTTSPSPTWPKRSTAARPSPSWPSTMPPCPPSSTSTSPSPPSKTSSPSTSSST- 286
Query: 613 VNATKAAASSDTPIGDEEKENETPKSPPRQNAPP--------------SPPSTHDAGSMP 658
+A K++ S T + P PP APP SPP T A S P
Sbjct: 287 TSAPKSSTRSTTAPPSSPPCSRPPAPPP---APPATSTSPTSRPARSASPPKTTTAPSTP 457
Query: 659 SPPHQGEKSCPGAATTSEAAQIEQAPAP 686
S + K CP + +S++A P P
Sbjct: 458 STSNPSRK-CPTTSPSSKSAPPLAPPTP 538
Score = 31.6 bits (70), Expect = 1.4
Identities = 23/55 (41%), Positives = 25/55 (44%)
Frame = +2
Query: 81 NRPIVRSNSPLHATHLPKTIMTSNLPHASNTRHPSSFSPSPIKTLLPHCHPFPRP 135
+RP RS SP T P T TSN T PSS S P+ P HP P P
Sbjct: 401 SRP-ARSASPPKTTTAPSTPSTSNPSRKCPTTSPSSKSAPPLAPPTPR-HPPPPP 559
>TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-protein
(AtAGP4) (AT5g10430/F12B17_220), partial (50%)
Length = 966
Score = 45.4 bits (106), Expect = 1e-04
Identities = 39/117 (33%), Positives = 46/117 (38%), Gaps = 3/117 (2%)
Frame = +2
Query: 537 APKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAEASV---AAATEPTAVPASSPASAG 593
AP + T S S P PTT A P A+ AA P A P +PA A
Sbjct: 206 APSQPPTTTPSPPPPRSAPAPAPTTPATPPPATPPPAATPPPAATPPPAATP--TPAPAP 379
Query: 594 ATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPS 650
TAA SS A+ S T + S +TP G P P +A P PPS
Sbjct: 380 PTAAPTPASSPAASPPSPSPTVTPSPTSPNTPPG--------PSPGPSGSAEPPPPS 526
Score = 41.6 bits (96), Expect = 0.001
Identities = 39/132 (29%), Positives = 50/132 (37%), Gaps = 8/132 (6%)
Frame = +2
Query: 555 PGAQP-----TTAAAPKGKNV-AEASVAAATEPTAVP--ASSPASAGATAAVAAESSIGA 606
PGA P TT + P ++ A A AT P A P A++P A A + A
Sbjct: 197 PGAAPSQPPTTTPSPPPPRSAPAPAPTTPATPPPATPPPAATPPPAATPPPAATPTPAPA 376
Query: 607 TAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEK 666
+A A+ AAS +P TP SP N PP P + P PP
Sbjct: 377 PPTAAPTPASSPAASPPSP-----SPTVTP-SPTSPNTPPGPSPGPSGSAEPPPPSAAFS 538
Query: 667 SCPGAATTSEAA 678
+ TS A
Sbjct: 539 ASKAFIATSALA 574
Score = 32.0 bits (71), Expect = 1.1
Identities = 25/96 (26%), Positives = 36/96 (37%)
Frame = +2
Query: 591 SAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPS 650
S T + + GA + ++ + P + T SPP + P+P
Sbjct: 95 STATTTTFSLDMGSGAVQLFLILGLLASSCLAQAPGAAPSQPPTTTPSPPPPRSAPAPAP 274
Query: 651 THDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAP 686
T A P+ P + P AAT AA APAP
Sbjct: 275 TTPATPPPATPPPA-ATPPPAATPPPAATPTPAPAP 379
>TC211828 similar to UP|RPIA_ARATH (Q9ZU38) Probable-ribose 5-phosphate
isomerase (Phosphoriboisomerase) , partial (47%)
Length = 729
Score = 45.4 bits (106), Expect = 1e-04
Identities = 40/150 (26%), Positives = 66/150 (43%), Gaps = 16/150 (10%)
Frame = +2
Query: 559 PTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATA-----ASAGV 613
PT ++P K + A++T P P+SSP + + SS + A A A +
Sbjct: 278 PTPISSPPRK--PPWTPASSTPPPPPPSSSPKTTSRKSPPTRPSSTSSPAWSSASAPAPL 451
Query: 614 NATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCP---- 669
+T + AS+ + + K + PP+ PSP ++ S P+PP + P
Sbjct: 452 PSTPSTASASSSAKENSKTSSESPPPPKPTTRPSPSASPSPISTPTPPSISPSTAPTRSI 631
Query: 670 GAATTSEAA-------QIEQAPAPEVGSSS 692
++T+S AA + +APA SSS
Sbjct: 632 PSSTSSRAAAAPSFEKRWSRAPARSSSSSS 721
>TC225559 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (47%)
Length = 870
Score = 45.1 bits (105), Expect = 1e-04
Identities = 45/149 (30%), Positives = 60/149 (40%), Gaps = 5/149 (3%)
Frame = +3
Query: 547 SSDAGTSNPGAQP---TTAAAPKGKNVAEASVAA--ATEPTAVPASSPASAGATAAVAAE 601
S A SN A P T A AP + A VA+ ++ P A PAS+PA++ T A
Sbjct: 138 SPAAAPSNTPATPAAATPAQAPSTVPKSPAPVASPKSSPPAATPASTPAASPTTTPAAPA 317
Query: 602 SSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPP 661
AAS A P+ P S P P + P+T A S P+P
Sbjct: 318 PVTKPPAASPPPATPPPATPPPAPVPVSSPPAPVPVSSPPAPVPVAAPTTPVAPS-PAPK 494
Query: 662 HQGEKSCPGAATTSEAAQIEQAPAPEVGS 690
H+ + GA S + PAP G+
Sbjct: 495 HKKKGKKHGAPAPS---PLLGPPAPPTGA 572
Score = 42.7 bits (99), Expect = 6e-04
Identities = 49/152 (32%), Positives = 61/152 (39%), Gaps = 7/152 (4%)
Frame = +3
Query: 531 QAEST--KAPKRRRLTKASSDAGT--SNPGAQPTT---AAAPKGKNVAEASVAAATEPTA 583
QA ST K+P K+S A T S P A PTT A AP K A AS AT P A
Sbjct: 195 QAPSTVPKSPAPVASPKSSPPAATPASTPAASPTTTPAAPAPVTKPPA-ASPPPATPPPA 371
Query: 584 VPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQN 643
P +P + A SS A A T A S P ++ + +P
Sbjct: 372 TPPPAPVPVSSPPAPVPVSSPPAPVPVAA--PTTPVAPSPAPKHKKKGKKHGAPAPSPLL 545
Query: 644 APPSPPSTHDAGSMPSPPHQGEKSCPGAATTS 675
PP+PP+ P P + S PG T +
Sbjct: 546 GPPAPPT-----GAPGPSE--DASSPGPGTAA 620
>TC225560 similar to UP|Q9FUL8 (Q9FUL8) Arabinogalactan protein, partial
(48%)
Length = 1030
Score = 45.1 bits (105), Expect = 1e-04
Identities = 40/145 (27%), Positives = 58/145 (39%), Gaps = 1/145 (0%)
Frame = +2
Query: 547 SSDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGA 606
S A SN A P A + + + A+ ++ PASSP A AT A + +S
Sbjct: 137 SPAAAPSNTPATPAAATPAQAPSTPNSPAPVASPKSSPPASSPKPATATPASSPAASPTT 316
Query: 607 T-AASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGE 665
T AA A AA+ P+ P S P P + P+T A + H+ +
Sbjct: 317 TPAAPAPATKPPAASPPPAPVPVSSPPAPVPVSSPPAPVPVAAPTTPVAPAPAPSKHKKK 496
Query: 666 KSCPGAATTSEAAQIEQAPAPEVGS 690
GA S + + PAP G+
Sbjct: 497 GKKHGAPAPSPS--LLGPPAPPTGA 565
Score = 42.0 bits (97), Expect = 0.001
Identities = 42/141 (29%), Positives = 57/141 (39%), Gaps = 2/141 (1%)
Frame = +2
Query: 537 APKRRRLTKASSDAG--TSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGA 594
+PK T ASS A T+ P A P A P + A V ++ P VP SSP A
Sbjct: 260 SPKPATATPASSPAASPTTTPAA-PAPATKPPAASPPPAPVPVSSPPAPVPVSSPP---A 427
Query: 595 TAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDA 654
VAA ++ A A + + K K++ P P PP+PP+
Sbjct: 428 PVPVAAPTTPVAPAPAPSKHKKKG------------KKHGAPAPSPSLLGPPAPPT---- 559
Query: 655 GSMPSPPHQGEKSCPGAATTS 675
P P + S PG AT +
Sbjct: 560 -GAPGPSE--DASSPGPATAA 613
Score = 33.9 bits (76), Expect = 0.29
Identities = 26/77 (33%), Positives = 32/77 (40%), Gaps = 1/77 (1%)
Frame = +2
Query: 611 AGVNA-TKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCP 669
AGV + AAA S+TP S P AP + P + S P P S P
Sbjct: 119 AGVGGQSPAAAPSNTPATPAAATPAQAPSTPNSPAPVASPKSSPPASSPKPATATPASSP 298
Query: 670 GAATTSEAAQIEQAPAP 686
A+ T+ A APAP
Sbjct: 299 AASPTTTPA----APAP 337
>AI444066 similar to GP|9294409|dbj polygalacturonase inhibitor-like protein
{Arabidopsis thaliana}, partial (27%)
Length = 336
Score = 44.3 bits (103), Expect = 2e-04
Identities = 38/111 (34%), Positives = 49/111 (43%), Gaps = 7/111 (6%)
Frame = +1
Query: 553 SNP-GAQPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASA 611
SNP PTTA + +G A+ A+ TA P +SP SA A S + A+
Sbjct: 22 SNPLSGSPTTAYSTRGPAPTAATTGTASPATATPTASPRSASAPVPSTPPSRSPSVLATC 201
Query: 612 GVNATKAAAS------SDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGS 656
V + + AS S +PIG E SP R A SPPS+ A S
Sbjct: 202 PVPSRRRFASSLTSPASSSPIGRE--------SPARSRA-ASPPSSSSASS 327
>TC219909 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (21%)
Length = 943
Score = 43.9 bits (102), Expect = 3e-04
Identities = 35/148 (23%), Positives = 63/148 (41%), Gaps = 5/148 (3%)
Frame = +1
Query: 559 PTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKA 618
P T+ +P +A+ + P P+ S S + A + ++G A S ++
Sbjct: 244 PKTSPSPSPSQSPKANPPTVSPPLPSPSPSVHSKSLSPASSPVPAVGTPAISPAISIPTL 423
Query: 619 AASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGSMP-----SPPHQGEKSCPGAAT 673
A + TP P P ++PPSP + + P SPP G S P A
Sbjct: 424 APETGTP---------PPSLGPSSSSPPSPGPSSSSPPSPGPSSSSPPPAGPTSPPSLAP 576
Query: 674 TSEAAQIEQAPAPEVGSSSYYNMLPNAI 701
+S + AP+P+ +S +++ P+++
Sbjct: 577 SSNST----APSPK---NSGFSVAPSSV 639
>TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partial (4%)
Length = 540
Score = 43.9 bits (102), Expect = 3e-04
Identities = 35/103 (33%), Positives = 53/103 (50%), Gaps = 10/103 (9%)
Frame = +1
Query: 531 QAESTKAPKRRRLTKASSDAGTSNPG---AQPTTAAAPKGKNVAEASVAAATEPTAVPAS 587
++ + AP + T SS T+ A TT++A + A+ AAAT A+
Sbjct: 4 ESAAATAPADAKPTATSSAVATTAAATSHAATTTSSAAAAAAASSAATAAATSSATTAAT 183
Query: 588 SPASAG----ATAAVAAES---SIGATAASAGVNATKAAASSD 623
+PA+A ATAA A+ S + ATAA++ V AT A A++D
Sbjct: 184 APAAAASATPATAAAASTSTTPTTTATAAASAVAATAATAATD 312
Score = 41.2 bits (95), Expect = 0.002
Identities = 29/81 (35%), Positives = 41/81 (49%), Gaps = 3/81 (3%)
Frame = +1
Query: 548 SDAGTSNPGAQPTTAAAPKGKNVAEASVAAATEPTAVPA---SSPASAGATAAVAAESSI 604
S A T+ A+PT ++ A S AA T +A A SS A+A AT++ ++
Sbjct: 7 SAAATAPADAKPTATSSAVATTAAATSHAATTTSSAAAAAAASSAATAAATSSATTAATA 186
Query: 605 GATAASAGVNATKAAASSDTP 625
A AASA AA++S TP
Sbjct: 187 PAAAASATPATAAAASTSTTP 249
Score = 36.2 bits (82), Expect = 0.058
Identities = 29/90 (32%), Positives = 42/90 (46%), Gaps = 5/90 (5%)
Frame = +1
Query: 528 AENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAE---ASVAAATEPTAV 584
A + A +T A +S A + + TAAA A A+ A+AT TA
Sbjct: 46 ATSSAVATTAAATSHAATTTSSAAAAAAASSAATAAATSSATTAATAPAAAASATPATAA 225
Query: 585 PASSPAS--AGATAAVAAESSIGATAASAG 612
AS+ + ATAA +A ++ ATAA+ G
Sbjct: 226 AASTSTTPTTTATAAASAVAATAATAATDG 315
Score = 33.5 bits (75), Expect = 0.38
Identities = 29/103 (28%), Positives = 41/103 (39%)
Frame = +1
Query: 576 AAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENET 635
AAAT P A A++ A A AA +S AT S+ A AAA+S T
Sbjct: 10 AAATAPA--DAKPTATSSAVATTAAATSHAATTTSS---AAAAAAASSAATAAATSSATT 174
Query: 636 PKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAA 678
+ P A +P + A + +P + A T+ A
Sbjct: 175 AATAPAAAASATPATAAAASTSTTPTTTATAAASAVAATAATA 303
Score = 33.1 bits (74), Expect = 0.49
Identities = 25/70 (35%), Positives = 34/70 (47%), Gaps = 5/70 (7%)
Frame = +1
Query: 546 ASSDAGTSNPGAQPTTAA-APKGKNVAEASVAAA----TEPTAVPASSPASAGATAAVAA 600
A+S A T+ + TTAA AP A + AAA T PT ++ ++ ATAA AA
Sbjct: 127 AASSAATAAATSSATTAATAPAAAASATPATAAAASTSTTPTTTATAAASAVAATAATAA 306
Query: 601 ESSIGATAAS 610
G +S
Sbjct: 307 TDGWGRNGSS 336
Score = 30.0 bits (66), Expect = 4.2
Identities = 25/76 (32%), Positives = 31/76 (39%), Gaps = 4/76 (5%)
Frame = -2
Query: 555 PGAQPTTAAAPKGKNVAEASVAAATEPTAVPA----SSPASAGATAAVAAESSIGATAAS 610
P A A + A A V E A A + A+AGA AAV AE A AA
Sbjct: 314 PSVAAVAAVAATAEAAAVAVVVGVVEVEAAAAVAGVAEAAAAGAVAAVVAEEVAAAVAAE 135
Query: 611 AGVNATKAAASSDTPI 626
A AAA+ + +
Sbjct: 134 ---EAAAAAAAEEVVV 96
>TC205436 homologue to UP|PRS6_ARATH (Q9SEI4) 26S protease regulatory subunit
6B homolog (26S proteasome AAA-ATPase subunit RPT3)
(Regulatory particle triple-A ATPase subunit 3), partial
(32%)
Length = 598
Score = 43.5 bits (101), Expect = 4e-04
Identities = 40/154 (25%), Positives = 70/154 (44%), Gaps = 5/154 (3%)
Frame = +3
Query: 530 NQAESTKAPKRRR---LTKASSDAGTSNPGAQPTTAAA--PKGKNVAEASVAAATEPTAV 584
N+ S + RR+ +KA+S A T P + T+ AA P +++ ++ +T T+
Sbjct: 114 NRRRSHRRSSRRQNPTTSKAASAAATPRPPTRTTSTAASRPCSASLSSSTSRKSTSRTSR 293
Query: 585 PASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPRQNA 644
SS +S+ + S +++AS+ +T+ AS D P + + S P +
Sbjct: 294 RISSASSSARRRRSSGSSRCRSSSASSWKWSTRTMASLDPP---PDPTTTSESSAPSTVS 464
Query: 645 PPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAA 678
SPP T + P+P P AA+ S A+
Sbjct: 465 FSSPPHTSRSTGTPTPSSTFFHRRPTAASRSSAS 566
>TC215397 weakly similar to UP|Q8S9J6 (Q8S9J6) AT5g10770/T30N20_40, partial
(76%)
Length = 1447
Score = 43.5 bits (101), Expect = 4e-04
Identities = 38/127 (29%), Positives = 57/127 (43%)
Frame = +1
Query: 573 ASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKE 632
A+ AA+ P P SS S+ A A A+SA A+ A+A++ +P ++
Sbjct: 394 ATSAASASP*PPPTSSTTSSSAAARTTR-------ASSAAPPASLASAATPSPSSNKLPP 552
Query: 633 NETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSS 692
N SP PP+PP+T S +PP P AAT+S P+P S+
Sbjct: 553 NTVKSSPTASPPPPAPPAT----SPSAPP-------PQAATSSTP---PSPPSPAAPPST 690
Query: 693 YYNMLPN 699
+LP+
Sbjct: 691 VSILLPS 711
Score = 37.7 bits (86), Expect = 0.020
Identities = 36/147 (24%), Positives = 57/147 (38%), Gaps = 18/147 (12%)
Frame = +1
Query: 532 AESTKAPKRRRLTKASSDAGTSN--PGAQPTTAAAPKGKNVAEASVAAATEPTAVPASSP 589
+ +T + R T+ASS A ++ A P+ ++ N ++S A+ P A PA+SP
Sbjct: 436 SSTTSSSAAARTTRASSAAPPASLASAATPSPSSNKLPPNTVKSSPTASPPPPAPPATSP 615
Query: 590 ASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGDEEKENETPKSPPR-------- 641
+ A AA SS + S + + +P ++ P SPP
Sbjct: 616 S---APPPQAATSSTPPSPPSPAAPPSTVSILLPSP*AVSNSQSPPPPSPPAVQSSTLAP 786
Query: 642 --------QNAPPSPPSTHDAGSMPSP 660
AP PPS + P P
Sbjct: 787 *SPASPQPPTAPSDPPSVKACQNTPPP 867
Score = 34.3 bits (77), Expect = 0.22
Identities = 44/154 (28%), Positives = 61/154 (39%), Gaps = 10/154 (6%)
Frame = +1
Query: 560 TTAAAPKGKNVAEASVAAATEPTAVPASSPASAGAT-AAVAAESSIGATAASAGVNATK- 617
+TA P + AS + PT+ SS A+A T A+ AA + A+AA+ ++ K
Sbjct: 367 STATVPSQLATSAASASP*PPPTSSTTSSSAAARTTRASSAAPPASLASAATPSPSSNKL 546
Query: 618 ---AAASSDTPIGDEEKENET-PKSPPRQNA----PPSPPSTHDAGSMPSPPHQGEKSCP 669
SS T T P +PP Q A PPSPPS +PP P
Sbjct: 547 PPNTVKSSPTASPPPPAPPATSPSAPPPQAATSSTPPSPPSP------AAPPSTVSILLP 708
Query: 670 GAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEP 703
S + P V SS+ P + +P
Sbjct: 709 SP*AVSNSQSPPPPSPPAVQSSTLAP*SPASPQP 810
Score = 32.7 bits (73), Expect = 0.65
Identities = 37/154 (24%), Positives = 58/154 (37%), Gaps = 6/154 (3%)
Frame = +1
Query: 558 QPTTAAAPKGKNVAEASVAAATEPTAVPASSPASAGA----TAAVAAESSIGATAASAGV 613
Q T P A + + AT P+ + A+S ASA T++ + S+ T ++
Sbjct: 313 QETIQDVPHPPRHAYTASSTATVPSQL-ATSAASASP*PPPTSSTTSSSAAARTTRASSA 489
Query: 614 NATKAAASSDTPIGDEEKENETPKSPPRQNAPPSPPSTHDAGS--MPSPPHQGEKSCPGA 671
+ AS+ TP SP PP+ + S P+PP + P
Sbjct: 490 APPASLASAATP------------SPSSNKLPPNTVKSSPTASPPPPAPPATSPSAPPPQ 633
Query: 672 ATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSE 705
A TS +PA + S P A+ S+
Sbjct: 634 AATSSTPPSPPSPAAPPSTVSILLPSP*AVSNSQ 735
>TC232974 weakly similar to GB|AAK54457.1|20384750|AY032998 DNA repair
protein XRCC3 {Arabidopsis thaliana;} , partial (40%)
Length = 909
Score = 43.1 bits (100), Expect = 5e-04
Identities = 36/124 (29%), Positives = 50/124 (40%)
Frame = +3
Query: 568 KNVAEASVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIG 627
+ + A + P AVP S AS A+ A + SS SA A + +++S++P
Sbjct: 6 RTYCNCNTAPRSAPWAVPYSIAASPAASPAPPSPSS------SARAVAARRSSASNSP-- 161
Query: 628 DEEKENETPKSPPRQNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPE 687
P SPP P PPS+ S PS P S A ++ AP P
Sbjct: 162 ------SPPSSPPLTAVSPPPPSS----STPSSPSPSAASATSHAPSAPPTPTSLAPTPA 311
Query: 688 VGSS 691
SS
Sbjct: 312 TASS 323
>BI969707
Length = 711
Score = 42.7 bits (99), Expect = 6e-04
Identities = 45/157 (28%), Positives = 67/157 (42%), Gaps = 6/157 (3%)
Frame = +3
Query: 525 RGGAENQAESTKAPKR--RRLTKASSDAGTSNPGAQPTTAAAPKGKNV-AEASVAAATEP 581
RG E ++ T R R+TK ++ + A T A N A A+ + A+
Sbjct: 174 RGDCEGESAKTA*YARFWGRITKGTTWSSPQKS*AHRCTFVARAAHNCSASATQSWASHC 353
Query: 582 TAVPASSPASAGATAAVAAESSIGATAASAGVNATKAAASSDTPIGD-EEKENETPKSPP 640
+A P P G G T+++A ++ +A TP E TP SPP
Sbjct: 354 SASPPPVPCDRGT----------GNTSSTA--SSPRAPPPRSTPSEPCSEPPPHTPPSPP 497
Query: 641 R--QNAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTS 675
R ++APPS + S P PP +G S P A++S
Sbjct: 498 RIYRSAPPSXMTPPPQSSTPXPPPRGPNSPPSTASSS 608
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.131 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,502,419
Number of Sequences: 63676
Number of extensions: 683401
Number of successful extensions: 10867
Number of sequences better than 10.0: 597
Number of HSP's better than 10.0 without gapping: 7492
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9596
length of query: 800
length of database: 12,639,632
effective HSP length: 105
effective length of query: 695
effective length of database: 5,953,652
effective search space: 4137788140
effective search space used: 4137788140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0315.1


