

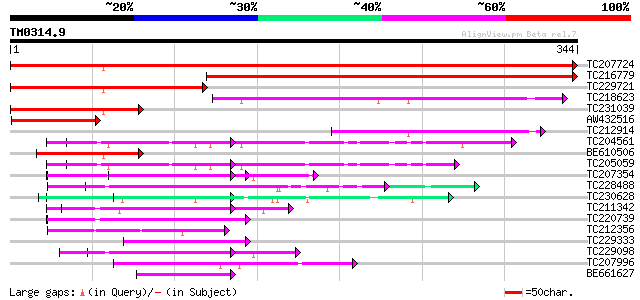
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0314.9
(344 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207724 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] ... 652 0.0
TC216779 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] ... 427 e-120
TC229721 similar to UP|Q9LMQ0 (Q9LMQ0) F7H2.18 protein, partial ... 230 6e-61
TC218623 homologue to PIR|T52386|T52386 mitotic checkpoint prote... 155 3e-38
TC231039 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] ... 140 1e-33
AW432516 weakly similar to PIR|A96839|A968 F23A5.2(form2) [impor... 84 7e-17
TC212914 69 4e-12
TC204561 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-... 65 4e-11
BE610506 similar to PIR|A96839|A968 F23A5.2(form2) [imported] - ... 64 8e-11
TC205059 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-... 62 3e-10
TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat prot... 62 5e-10
TC228488 similar to UP|Q8LF96 (Q8LF96) PRL1 protein, partial (70%) 60 1e-09
TC230628 homologue to UP|Q9XIJ3 (Q9XIJ3) T10O24.21, partial (41%) 58 7e-09
TC211342 similar to UP|Q9LVF2 (Q9LVF2) Arabidopsis thaliana geno... 57 1e-08
TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nu... 56 2e-08
TC212356 similar to UP|Q9FG99 (Q9FG99) Similarity to GTP-binding... 56 3e-08
TC229333 similar to UP|Q6H976 (Q6H976) STYLOSA protein, partial ... 55 6e-08
TC229098 similar to PIR|T46032|T46032 WD-40 repeat regulatory pr... 54 8e-08
TC207996 UP|Q6T2Z5 (Q6T2Z5) WD-repeat cell cycle regulatory prot... 54 1e-07
BE661627 similar to GP|2443881|gb contains beta-transducin motif... 53 2e-07
>TC207724 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(98%)
Length = 1343
Score = 652 bits (1682), Expect = 0.0
Identities = 302/347 (87%), Positives = 323/347 (93%), Gaps = 3/347 (0%)
Frame = +2
Query: 1 MSTFLSNTNQNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARN---VA 57
MSTF + N NPNKS+EV QPPSDS+SS+CFSPKAN LVATSWDNQVRCWE+ RN V
Sbjct: 71 MSTFGAAANTNPNKSYEVAQPPSDSISSICFSPKANFLVATSWDNQVRCWEITRNGTVVN 250
Query: 58 TAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAW 117
+ PKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPL+SGGQPMTVAMHDAPVKDIAW
Sbjct: 251 STPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLMSGGQPMTVAMHDAPVKDIAW 430
Query: 118 IPEMSLLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNL 177
IPEM+LL TGSWD+T+KYWDTRQ NPVHTQQLP+RCYA++VKHPLMVVGTADRN++V+NL
Sbjct: 431 IPEMNLLATGSWDKTLKYWDTRQSNPVHTQQLPDRCYAITVKHPLMVVGTADRNLIVFNL 610
Query: 178 QNPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHRE 237
QNPQ E+KRI+SPLKYQTR +AAFPDQQGFLVGSIEGRVGVHHLDD+QQ KNFTFKCHRE
Sbjct: 611 QNPQTEYKRIVSPLKYQTRSVAAFPDQQGFLVGSIEGRVGVHHLDDAQQNKNFTFKCHRE 790
Query: 238 GNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSI 297
NEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAM RCSQPIPCS FNNDGSI
Sbjct: 791 NNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMQRCSQPIPCSTFNNDGSI 970
Query: 298 FAYSVCYDWSKGAENHNPTTAKTNIYLHLPQESEVKGKPRIGATGRK 344
FAY+VCYDWSKGAENHNP TAK IYLHLPQESEVKGKPR GATGRK
Sbjct: 971 FAYAVCYDWSKGAENHNPATAKNYIYLHLPQESEVKGKPRAGATGRK 1111
>TC216779 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(60%)
Length = 1218
Score = 427 bits (1099), Expect = e-120
Identities = 200/225 (88%), Positives = 211/225 (92%)
Frame = +3
Query: 120 EMSLLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQN 179
+ S+L T + T +YWDTRQ NPVHTQQLPERCYAM+V+HPLMVVGTADRN++VYNLQN
Sbjct: 108 DASVLST*VFLATFRYWDTRQSNPVHTQQLPERCYAMTVRHPLMVVGTADRNLIVYNLQN 287
Query: 180 PQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGN 239
PQVEFKRI+SPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQ GKNFTFKCHREGN
Sbjct: 288 PQVEFKRIVSPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQHGKNFTFKCHREGN 467
Query: 240 EIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIFA 299
EIYSVNSLNFHPVHHTFAT+GSDGAFNFWDKDSKQRLKAMLRCSQPIPCS FNNDGSIFA
Sbjct: 468 EIYSVNSLNFHPVHHTFATSGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSTFNNDGSIFA 647
Query: 300 YSVCYDWSKGAENHNPTTAKTNIYLHLPQESEVKGKPRIGATGRK 344
YSVCYDWSKGAEN NP AKT I+LHLPQESEVKGKPRIGATGRK
Sbjct: 648 YSVCYDWSKGAENSNPAAAKTYIFLHLPQESEVKGKPRIGATGRK 782
Score = 30.4 bits (67), Expect = 1.2
Identities = 19/78 (24%), Positives = 38/78 (48%), Gaps = 2/78 (2%)
Frame = +3
Query: 5 LSNTNQNPNKSFEVNQPPSD--SVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKA 62
L ++ N +F+ ++ ++ SV+SL F P + + D W+ ++ KA
Sbjct: 411 LDDSQHGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATSGSDGAFNFWD--KDSKQRLKA 584
Query: 63 SISHDQPVLCSAWKDDGT 80
+ QP+ CS + +DG+
Sbjct: 585 MLRCSQPIPCSTFNNDGS 638
>TC229721 similar to UP|Q9LMQ0 (Q9LMQ0) F7H2.18 protein, partial (81%)
Length = 1121
Score = 230 bits (587), Expect = 6e-61
Identities = 108/123 (87%), Positives = 113/123 (91%), Gaps = 3/123 (2%)
Frame = +1
Query: 1 MSTFLSNTNQNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARN---VA 57
MS FLSNTN NPNKSFEVNQPP+DSVSSL FSPKAN LVATSWDNQVRCWEVARN VA
Sbjct: 58 MSNFLSNTNPNPNKSFEVNQPPTDSVSSLSFSPKANFLVATSWDNQVRCWEVARNGVNVA 237
Query: 58 TAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAW 117
T PKASI+HD PVLCS WKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAP+K++AW
Sbjct: 238 TVPKASITHDHPVLCSTWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPIKELAW 417
Query: 118 IPE 120
IPE
Sbjct: 418 IPE 426
Score = 29.6 bits (65), Expect = 2.1
Identities = 15/65 (23%), Positives = 34/65 (52%)
Frame = +1
Query: 112 VKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRN 171
V +++ P+ + LV SWD ++ W+ + N V+ +P+ +++ HP++ D
Sbjct: 133 VSSLSFSPKANFLVATSWDNQVRCWEVAR-NGVNVATVPKA--SITHDHPVLCSTWKDDG 303
Query: 172 ILVYN 176
V++
Sbjct: 304 TTVFS 318
Score = 29.6 bits (65), Expect = 2.1
Identities = 12/24 (50%), Positives = 18/24 (75%)
Frame = +2
Query: 111 PVKDIAWIPEMSLLVTGSWDRTIK 134
P K++ + +LLVTGSWD+T+K
Sbjct: 398 PSKNLLGFLKXNLLVTGSWDKTMK 469
>TC218623 homologue to PIR|T52386|T52386 mitotic checkpoint protein
[imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (67%)
Length = 904
Score = 155 bits (392), Expect = 3e-38
Identities = 82/226 (36%), Positives = 130/226 (57%), Gaps = 11/226 (4%)
Frame = +3
Query: 124 LVTGSWDRTIKYWDTR-----QPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQ 178
L+TGSWD+T+K WD R + V T PER Y++S+ +VV TA R++ +Y+L+
Sbjct: 9 LITGSWDKTLKCWDPRGASGQERTLVGTYPQPERVYSLSLVGHRLVVATAGRHVNIYDLR 188
Query: 179 NPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLD--DSQQGKNFTFKCHR 236
N +R S LKYQTRC+ +P+ G+ + S+EGRV + D ++ Q K + FKCHR
Sbjct: 189 NMSQPEQRRESSLKYQTRCVHCYPNGTGYALSSVEGRVAMEFFDLSEASQAKKYAFKCHR 368
Query: 237 EGNE----IYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFN 292
+ +Y VN++ FHP++ TFAT G DG N WD ++K+RL + + +F+
Sbjct: 369 KSEAGRDIVYPVNAIAFHPIYGTFATGGCDGYVNVWDGNNKKRLYQYSKYPTSVAALSFS 548
Query: 293 NDGSIFAYSVCYDWSKGAENHNPTTAKTNIYLHLPQESEVKGKPRI 338
DG + A + Y + +G ++ + I++ E EVK KP++
Sbjct: 549 RDGRLLAVASSYTFEEGPKSQE----QDAIFVRSVNEIEVKPKPKV 674
>TC231039 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(23%)
Length = 541
Score = 140 bits (352), Expect = 1e-33
Identities = 66/84 (78%), Positives = 72/84 (85%), Gaps = 3/84 (3%)
Frame = +2
Query: 1 MSTFLSNTNQNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARN---VA 57
MSTF + N NPNKS+EV QPPSDS+SSLCFSPKAN LVATSWDNQVRCWE+ RN V
Sbjct: 290 MSTFGAAANTNPNKSYEVAQPPSDSISSLCFSPKANFLVATSWDNQVRCWEITRNGTVVN 469
Query: 58 TAPKASISHDQPVLCSAWKDDGTT 81
+ PKASISH+QPVLCSAWKDDGTT
Sbjct: 470 STPKASISHEQPVLCSAWKDDGTT 541
>AW432516 weakly similar to PIR|A96839|A968 F23A5.2(form2) [imported] -
Arabidopsis thaliana, partial (14%)
Length = 169
Score = 84.3 bits (207), Expect = 7e-17
Identities = 36/54 (66%), Positives = 45/54 (82%)
Frame = +2
Query: 2 STFLSNTNQNPNKSFEVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARN 55
STF ++ N N NKS+EV+QPPSD++SS+ FSPK N+ V TSWDNQVRCWE+ RN
Sbjct: 2 STFGADANANANKSYEVSQPPSDAISSIRFSPKDNVFVTTSWDNQVRCWEITRN 163
>TC212914
Length = 856
Score = 68.6 bits (166), Expect = 4e-12
Identities = 39/136 (28%), Positives = 69/136 (50%), Gaps = 6/136 (4%)
Frame = +3
Query: 196 RCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGK-NFTFKCHREGNE----IYSVNSLNFH 250
RC+++ PD +GF VGS++GRV + S + + F+CH + + + SVN + F
Sbjct: 162 RCVSSIPDAEGFAVGSVDGRVSLQISYPSGSDEIRYIFRCHPKSKDGRHYLVSVNDIAFS 341
Query: 251 P-VHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDGSIFAYSVCYDWSKG 309
P V FAT ++G WD S++RL + R + ++N+ G + A + + + +
Sbjct: 342 PLVSGAFATGDNEGYVTIWDAGSRRRLVELPRYPNSVASLSYNHTGQLLAVASSHTYQEA 521
Query: 310 AENHNPTTAKTNIYLH 325
E P I++H
Sbjct: 522 KEIEKP----PRIFIH 557
>TC204561 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-binding
protein beta subunit-like protein, complete
Length = 1419
Score = 65.1 bits (157), Expect = 4e-11
Identities = 68/286 (23%), Positives = 118/286 (40%), Gaps = 13/286 (4%)
Frame = +1
Query: 35 ANLLVATSWDNQVRCWEVARNVAT--APKASIS-HDQPVLCSAWKDDGTTVFSGGCDKQV 91
++++V S D + W + + T P+ ++ H V DG SG D ++
Sbjct: 226 SDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSGSWDGEL 405
Query: 92 KMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTR-------QPNPV 144
++W L +G H V +A+ + +V+ S DRTIK W+T Q
Sbjct: 406 RLWDLAAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDGDA 585
Query: 145 HTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAFPDQ 204
H+ + ++ S P +V + DR + V+NL N + + ++ +A PD
Sbjct: 586 HSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTN--CKLRNTLAGHNGYVNTVAVSPD- 756
Query: 205 QGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGA 264
G L S G+ GV L D +GK + I +++L F P + + A ++ +
Sbjct: 757 -GSLCAS-GGKDGVILLWDLAEGKRL---YSLDAGSI--IHALCFSP-NRYWLCAATEQS 912
Query: 265 FNFWDKDSK---QRLKAMLRCSQPIPCSAFNNDGSIFAYSVCYDWS 307
WD +SK + LK L+ N + Y +WS
Sbjct: 913 IKIWDLESKSIVEDLKVDLKTEADATSGGGNANKKKVIYCTSLNWS 1050
Score = 54.7 bits (130), Expect = 6e-08
Identities = 34/120 (28%), Positives = 57/120 (47%), Gaps = 5/120 (4%)
Frame = +1
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S V + S ++ SWD ++R W++A T+ + + H + VL A+ D +
Sbjct: 331 SHFVQDVVLSSDGQFALSGSWDGELRLWDLA--AGTSARRFVGHTKDVLSVAFSIDNRQI 504
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAP---VKDIAWIPE--MSLLVTGSWDRTIKYWD 137
S D+ +K+W L G T+ DA V + + P +V+ SWDRT+K W+
Sbjct: 505 VSASRDRTIKLWNTL-GECKYTIQDGDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWN 681
Score = 35.4 bits (80), Expect = 0.038
Identities = 21/87 (24%), Positives = 41/87 (46%), Gaps = 16/87 (18%)
Frame = +1
Query: 26 VSSLCFSPKANLLVATSWDNQVRCWEVARNV--------------ATAPKASISHDQPVL 71
+ +LCFSP L A + + ++ W++ AT+ + + + +
Sbjct: 853 IHALCFSPNRYWLCAAT-EQSIKIWDLESKSIVEDLKVDLKTEADATSGGGNANKKKVIY 1029
Query: 72 CSA--WKDDGTTVFSGGCDKQVKMWPL 96
C++ W DG+T+FSG D V++W +
Sbjct: 1030CTSLNWSADGSTLFSGYTDGVVRVWAI 1110
>BE610506 similar to PIR|A96839|A968 F23A5.2(form2) [imported] -
Arabidopsis thaliana, partial (5%)
Length = 205
Score = 64.3 bits (155), Expect = 8e-11
Identities = 31/68 (45%), Positives = 43/68 (62%), Gaps = 3/68 (4%)
Frame = +2
Query: 17 EVNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARN---VATAPKASISHDQPVLCS 73
+V QPPSD +C + KA+ LVAT W+NQVRCWE+ RN + P A + ++ PV S
Sbjct: 2 KVAQPPSDPKRRVCINAKASFLVATLWNNQVRCWEITRNGTEATSTPNAWMEYELPVGSS 181
Query: 74 AWKDDGTT 81
+D+G T
Sbjct: 182PPQDEGPT 205
>TC205059 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-binding
protein beta subunit-like protein, complete
Length = 1492
Score = 62.4 bits (150), Expect = 3e-10
Identities = 61/249 (24%), Positives = 107/249 (42%), Gaps = 10/249 (4%)
Frame = +2
Query: 35 ANLLVATSWDNQVRCWEVARNVAT--APKASIS-HDQPVLCSAWKDDGTTVFSGGCDKQV 91
++++V S D + W + + T P+ ++ H V DG SG D ++
Sbjct: 302 SDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSGSWDGEL 481
Query: 92 KMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTR-------QPNPV 144
++W L +G H V +A+ + +V+ S DRTIK W+T Q
Sbjct: 482 RLWDLAAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDGDA 661
Query: 145 HTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQNPQVEFKRIISPLKYQTRCLAAFPDQ 204
H+ + ++ S P +V + DR + V+NL N + + ++ +A PD
Sbjct: 662 HSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTN--CKLRNTLAGHNGYVNTVAVSPD- 832
Query: 205 QGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGA 264
G L S G+ GV L D +GK + I +++L F P + + A ++ +
Sbjct: 833 -GSLCAS-GGKDGVILLWDLAEGKRL---YSLDAGSI--IHALCFSP-NRYWLCAATEQS 988
Query: 265 FNFWDKDSK 273
WD +SK
Sbjct: 989 IKIWDLESK 1015
Score = 54.7 bits (130), Expect = 6e-08
Identities = 34/120 (28%), Positives = 57/120 (47%), Gaps = 5/120 (4%)
Frame = +2
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S V + S ++ SWD ++R W++A T+ + + H + VL A+ D +
Sbjct: 407 SHFVQDVVLSSDGQFALSGSWDGELRLWDLA--AGTSARRFVGHTKDVLSVAFSIDNRQI 580
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAP---VKDIAWIPE--MSLLVTGSWDRTIKYWD 137
S D+ +K+W L G T+ DA V + + P +V+ SWDRT+K W+
Sbjct: 581 VSASRDRTIKLWNTL-GECKYTIQDGDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWN 757
Score = 28.5 bits (62), Expect = 4.6
Identities = 10/20 (50%), Positives = 14/20 (70%)
Frame = +3
Query: 75 WKDDGTTVFSGGCDKQVKMW 94
W DG+T+FSG D V++W
Sbjct: 1122 WSSDGSTLFSGYTDGVVRVW 1181
>TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat protein 5,
partial (57%)
Length = 1124
Score = 61.6 bits (148), Expect = 5e-10
Identities = 37/124 (29%), Positives = 59/124 (46%), Gaps = 1/124 (0%)
Frame = +1
Query: 24 DSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVF 83
++VS + FS LL + S D + W A T + H + + AW D +
Sbjct: 139 NAVSCVKFSNDGTLLASASLDKTLIIWSSA--TLTLCHRLVGHSEGISDLAWSSDSHYIC 312
Query: 84 SGGCDKQVKMWPLLSGGQPMTVAM-HDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPN 142
S D+ +++W GG + + HD V + + P+ S +V+GS+D TIK WD +
Sbjct: 313 SASDDRTLRIWDATVGGGCIKILRGHDDAVFCVNFNPQSSYIVSGSFDETIKVWDVKTGK 492
Query: 143 PVHT 146
VHT
Sbjct: 493 CVHT 504
Score = 60.8 bits (146), Expect = 8e-10
Identities = 41/169 (24%), Positives = 79/169 (46%), Gaps = 4/169 (2%)
Frame = +1
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S+ +S L +S ++ + + S D +R W+ K HD V C + + +
Sbjct: 262 SEGISDLAWSSDSHYICSASDDRTLRIWDATVGGGCI-KILRGHDDAVFCVNFNPQSSYI 438
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPN 142
SG D+ +K+W + +G T+ H PV + + + +L+++ S D + K WDT N
Sbjct: 439 VSGSFDETIKVWDVKTGKCVHTIKGHTMPVTSVHYNRDGNLIISASHDGSCKIWDTETGN 618
Query: 143 PVHT---QQLPERCYA-MSVKHPLMVVGTADRNILVYNLQNPQVEFKRI 187
+ T + P +A S L++ T + + ++N + +V FK +
Sbjct: 619 LLKTLIEDKAPAVSFAKFSPNGKLILAATLNDTLKLWNYGSGEV-FKNL 762
Score = 45.1 bits (105), Expect = 5e-05
Identities = 21/77 (27%), Positives = 36/77 (46%)
Frame = +1
Query: 61 KASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPE 120
K H+ V C + +DGT + S DK + +W + + H + D+AW +
Sbjct: 118 KTLTDHENAVSCVKFSNDGTLLASASLDKTLIIWSSATLTLCHRLVGHSEGISDLAWSSD 297
Query: 121 MSLLVTGSWDRTIKYWD 137
+ + S DRT++ WD
Sbjct: 298 SHYICSASDDRTLRIWD 348
>TC228488 similar to UP|Q8LF96 (Q8LF96) PRL1 protein, partial (70%)
Length = 1303
Score = 60.1 bits (144), Expect = 1e-09
Identities = 57/211 (27%), Positives = 91/211 (43%), Gaps = 4/211 (1%)
Frame = +3
Query: 24 DSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVF 83
+ V L S + + + D QV+CW++ +N ++ H V C A +
Sbjct: 639 EQVRGLAVSNRHTYMFSAGDDKQVKCWDLEQNKVI--RSYHGHLSGVYCLALHPTIDVLL 812
Query: 84 SGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNP 143
+GG D ++W + S Q ++ HD V + P +VTGS D TIK WD R
Sbjct: 813 TGGRDSVCRVWDIRSKMQIHALSGHDNTVCSVFTRPTDPQVVTGSHDTTIKMWDLRYGKT 992
Query: 144 VHTQQLPERCYAMSVKHP---LMVVGTADRNILVYNLQNPQVEF-KRIISPLKYQTRCLA 199
+ T ++ +HP +AD NI +NL P+ EF ++S K +A
Sbjct: 993 MSTLTNHKKSVRAMAQHPKEQAFASASAD-NIKKFNL--PKGEFCHNMLSQQKTIINAMA 1163
Query: 200 AFPDQQGFLVGSIEGRVGVHHLDDSQQGKNF 230
+++G +V G G D + G NF
Sbjct: 1164V--NEEGVMVTG--GDNGSMWFWDWKSGHNF 1244
Score = 55.1 bits (131), Expect = 5e-08
Identities = 58/245 (23%), Positives = 96/245 (38%), Gaps = 6/245 (2%)
Frame = +3
Query: 47 VRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVA 106
++ W++A V H + V A + T +FS G DKQVK W L +
Sbjct: 582 IKIWDLASGVLKLTLTG--HIEQVRGLAVSNRHTYMFSAGDDKQVKCWDLEQNKVIRSYH 755
Query: 107 MHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSVKHPL---M 163
H + V +A P + +L+TG D + WD R +H + P +
Sbjct: 756 GHLSGVYCLALHPTIDVLLTGGRDSVCRVWDIRSKMQIHALSGHDNTVCSVFTRPTDPQV 935
Query: 164 VVGTADRNILVYNLQNPQVEFKRIISPL---KYQTRCLAAFPDQQGFLVGSIEGRVGVHH 220
V G+ D I +++L+ + + +S L K R +A P +Q F S + + +
Sbjct: 936 VTGSHDTTIKMWDLR-----YGKTMSTLTNHKKSVRAMAQHPKEQAFASASAD-NIKKFN 1097
Query: 221 LDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAML 280
L + F + I + ++N V T G +G+ FWD S +
Sbjct: 1098LPKGE----FCHNMLSQQKTIINAMAVNEEGV---MVTGGDNGSMWFWDWKSGHNFQQSQ 1256
Query: 281 RCSQP 285
QP
Sbjct: 1257TIVQP 1271
>TC230628 homologue to UP|Q9XIJ3 (Q9XIJ3) T10O24.21, partial (41%)
Length = 1119
Score = 57.8 bits (138), Expect = 7e-09
Identities = 58/255 (22%), Positives = 100/255 (38%), Gaps = 8/255 (3%)
Frame = +2
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S +V +CFS ++ +D ++ W+ + A+ V + +D +
Sbjct: 20 SKAVRDICFSNDGTKFLSAGYDKNIKYWDTETGQVISTFATGKIPYVVKLNPDEDKQNVL 199
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPN 142
+G DK++ W + +G H V I ++ VT S D++++ W+ P
Sbjct: 200 LAGMSDKKIVQWDMNTGQITQEYDQHLGAVNTITFVDNNRRFVTSSDDKSLRVWEFSIPV 379
Query: 143 PVHTQQLPERCYAMSVK-HP---LMVVGTADRNILVYNLQN--PQVEFKRIISPLKYQTR 196
+ P S+ HP + + D IL+Y+ + + KR +
Sbjct: 380 VIKYISEPHMHSMPSISLHPNANWLAAQSLDNQILIYSTREKFQLNKRKRFGGHIVAGYA 559
Query: 197 CLAAF-PDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHT 255
C F PD Q + G EG+ + + T KCH EG I +HP+ +
Sbjct: 560 CQVNFSPDGQYVMSGDGEGKCWFWDWKTCKVYR--TLKCH-EGVCI----GCEWHPLEQS 718
Query: 256 -FATAGSDGAFNFWD 269
AT G DG +WD
Sbjct: 719 KVATCGWDGMIKYWD 763
Score = 45.4 bits (106), Expect = 4e-05
Identities = 33/124 (26%), Positives = 48/124 (38%), Gaps = 4/124 (3%)
Frame = +2
Query: 18 VNQPPSDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHD--QPVLCSA- 74
+++P S+ S+ P AN L A S DNQ+ + + C
Sbjct: 392 ISEPHMHSMPSISLHPNANWLAAQSLDNQILIYSTREKFQLNKRKRFGGHIVAGYACQVN 571
Query: 75 WKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIP-EMSLLVTGSWDRTI 133
+ DG V SG + + W + T+ H+ W P E S + T WD I
Sbjct: 572 FSPDGQYVMSGDGEGKCWFWDWKTCKVYRTLKCHEGVCIGCEWHPLEQSKVATCGWDGMI 751
Query: 134 KYWD 137
KYWD
Sbjct: 752 KYWD 763
Score = 43.5 bits (101), Expect = 1e-04
Identities = 49/215 (22%), Positives = 85/215 (38%), Gaps = 9/215 (4%)
Frame = +2
Query: 64 ISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAP--VKDIAWIPEM 121
+ H + V + +DGT S G DK +K W +G T A P VK +
Sbjct: 11 MGHSKAVRDICFSNDGTKFLSAGYDKNIKYWDTETGQVISTFATGKIPYVVKLNPDEDKQ 190
Query: 122 SLLVTGSWDRTIKYWDTRQPNPVHTQQLPERCYAMSV-----KHPLMVVGTADRNILVYN 176
++L+ G D+ I WD TQ+ + A++ + V + D+++ V+
Sbjct: 191 NVLLAGMSDKKIVQWDMNTGQ--ITQEYDQHLGAVNTITFVDNNRRFVTSSDDKSLRVWE 364
Query: 177 LQNPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHR 236
P V K I P + ++ P+ S++ ++ ++ S + K K R
Sbjct: 365 FSIP-VVIKYISEPHMHSMPSISLHPNANWLAAQSLDNQILIY----STREKFQLNKRKR 529
Query: 237 EGNEIYS--VNSLNFHPVHHTFATAGSDGAFNFWD 269
G I + +NF P + +G FWD
Sbjct: 530 FGGHIVAGYACQVNFSPDGQYVMSGDGEGKCWFWD 634
>TC211342 similar to UP|Q9LVF2 (Q9LVF2) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MIL23, partial (29%)
Length = 862
Score = 57.0 bits (136), Expect = 1e-08
Identities = 41/156 (26%), Positives = 69/156 (43%), Gaps = 6/156 (3%)
Frame = +1
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASIS---HDQPVLCSAWKDDG 79
+D + SP A + D+ V+ + A K +S H PVLC DG
Sbjct: 220 NDDALVVAISPDAKYIAVALLDSTVKV-----HFADTFKFFLSLYGHKLPVLCMDISSDG 384
Query: 80 TTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTR 139
+ +G DK +K+W L G ++ H V + ++P+ + + DR +KYWD
Sbjct: 385 DLIVTGSADKNIKIWGLDFGDCHKSIFAHADSVMAVQFVPKTHYVFSVGKDRLVKYWDAD 564
Query: 140 QPNPVHTQQLPER---CYAMSVKHPLMVVGTADRNI 172
+ + T + C A+S + +V G+ DR+I
Sbjct: 565 KFELLLTLEGHHADIWCLAVSNRGDFIVTGSHDRSI 672
Score = 43.9 bits (102), Expect = 1e-04
Identities = 30/106 (28%), Positives = 48/106 (44%)
Frame = +1
Query: 32 SPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQV 91
S +L+V S D ++ W + + K+ +H V+ + VFS G D+ V
Sbjct: 373 SSDGDLIVTGSADKNIKIWGL--DFGDCHKSIFAHADSVMAVQFVPKTHYVFSVGKDRLV 546
Query: 92 KMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWD 137
K W +T+ H A + +A +VTGS DR+I+ WD
Sbjct: 547 KYWDADKFELLLTLEGHHADIWCLAVSNRGDFIVTGSHDRSIRRWD 684
Score = 33.1 bits (74), Expect = 0.19
Identities = 47/212 (22%), Positives = 75/212 (35%), Gaps = 20/212 (9%)
Frame = +1
Query: 104 TVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNPV-----HTQQLPERCYAMSV 158
T+ H V + + SLL +GS D + WD + H Q ++ +V
Sbjct: 25 TLNGHKGAVTTLRYNKAGSLLASGSRDNDVILWDVVGETGLFRLRGHRDQAAKQLTVSNV 204
Query: 159 K------HPLMVVGTADRNILVYNLQNPQVE------FKRIISPL--KYQTRCLAAFPDQ 204
L+V + D + L + V+ FK +S K C+ D
Sbjct: 205 STMKMNDDALVVAISPDAKYIAVALLDSTVKVHFADTFKFFLSLYGHKLPVLCMDISSDG 384
Query: 205 QGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHRE-GNEIYSVNSLNFHPVHHTFATAGSDG 263
+ GS + + + LD CH+ SV ++ F P H + G D
Sbjct: 385 DLIVTGSADKNIKIWGLDFGD--------CHKSIFAHADSVMAVQFVPKTHYVFSVGKDR 540
Query: 264 AFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDG 295
+WD D + L + I C A +N G
Sbjct: 541 LVKYWDADKFELLLTLEGHHADIWCLAVSNRG 636
>TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nuclear
ribonucleoprotein Prp4 (U4/U6 snRNP 60 kDa protein) (WD
splicing factor Prp4) (hPrp4), partial (34%)
Length = 1206
Score = 56.2 bits (134), Expect = 2e-08
Identities = 33/123 (26%), Positives = 54/123 (43%)
Frame = +1
Query: 24 DSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVF 83
D ++ + F P L S+D R W++ + H + V A+ +DG+
Sbjct: 322 DRLARIAFHPSGKYLGTASFDKTWRLWDIETGDELLLQEG--HSRSVYGLAFHNDGSLAA 495
Query: 84 SGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNP 143
S G D ++W L +G + + H PV I++ P L TG D T + WD R+
Sbjct: 496 SCGLDSLARVWDLRTGRSILALEGHVKPVLGISFSPNGYHLATGGEDNTCRIWDLRKKKS 675
Query: 144 VHT 146
+T
Sbjct: 676 FYT 684
Score = 51.6 bits (122), Expect = 5e-07
Identities = 36/125 (28%), Positives = 55/125 (43%), Gaps = 1/125 (0%)
Frame = +1
Query: 23 SDSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTV 82
S SV L F +L + D+ R W++ + A H +PVL ++ +G +
Sbjct: 445 SRSVYGLAFHNDGSLAASCGLDSLARVWDL--RTGRSILALEGHVKPVLGISFSPNGYHL 618
Query: 83 FSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIP-EMSLLVTGSWDRTIKYWDTRQP 141
+GG D ++W L T+ H + + + P E LVT S+D T K W R
Sbjct: 619 ATGGEDNTCRIWDLRKKKSFYTIPAHSNLISQVKFEPHEGYFLVTASYDMTAKVWSGRDF 798
Query: 142 NPVHT 146
PV T
Sbjct: 799 KPVKT 813
Score = 38.5 bits (88), Expect = 0.004
Identities = 33/126 (26%), Positives = 51/126 (40%), Gaps = 2/126 (1%)
Frame = +1
Query: 26 VSSLCFSPKANLLVATSWDNQVRCWEVARNVA--TAPKASISHDQPVLCSAWKDDGTTVF 83
V + FSP L DN R W++ + + T P S Q +G +
Sbjct: 580 VLGISFSPNGYHLATGGEDNTCRIWDLRKKKSFYTIPAHSNLISQ---VKFEPHEGYFLV 750
Query: 84 SGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNP 143
+ D K+W T++ H+A V + + + +VT S DRTIK W + NP
Sbjct: 751 TASYDMTAKVWSGRDFKPVKTLSGHEAKVTSVDVLGDGGSIVTVSHDRTIKLWSS---NP 921
Query: 144 VHTQQL 149
Q +
Sbjct: 922 TDEQAM 939
Score = 37.4 bits (85), Expect = 0.010
Identities = 54/250 (21%), Positives = 95/250 (37%), Gaps = 6/250 (2%)
Frame = +1
Query: 62 ASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEM 121
+ I D+P+ ++ DG + + K+W + + H D+A+ P
Sbjct: 58 SEIGDDRPLSGCSFSRDGKWLATCSLTGASKLWSMPKIKKHSIFKGHTERATDVAYSPVH 237
Query: 122 SLLVTGSWDRTIKYWD---TRQPNPVHTQQLPERCYAMSVKHPLMVVGTADRNILVYNLQ 178
L T S DRT KYW+ + H +L + S K+ +GTA + + L
Sbjct: 238 DHLATASADRTAKYWNQGSLLKTFEGHLDRLARIAFHPSGKY----LGTASFD-KTWRLW 402
Query: 179 NPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGSIEGRVGVHHLD---DSQQGKNFTFKCH 235
+ + + ++ ++ AF + GS+ G+ L D + G++
Sbjct: 403 DIETGDELLLQEGHSRSVYGLAFHND-----GSLAASCGLDSLARVWDLRTGRSI---LA 558
Query: 236 REGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDG 295
EG+ + V ++F P + AT G D WD K+ + S I F
Sbjct: 559 LEGH-VKPVLGISFSPNGYHLATGGEDNTCRIWDLRKKKSFYTIPAHSNLISQVKFEPHE 735
Query: 296 SIFAYSVCYD 305
F + YD
Sbjct: 736 GYFLVTASYD 765
>TC212356 similar to UP|Q9FG99 (Q9FG99) Similarity to GTP-binding regulatory
protein and WD-repeat protein, partial (60%)
Length = 1044
Score = 55.8 bits (133), Expect = 3e-08
Identities = 33/114 (28%), Positives = 58/114 (49%), Gaps = 4/114 (3%)
Frame = +1
Query: 24 DSVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVF 83
D+VSSL S L + SWD ++ W ++ A +HD + A DG V+
Sbjct: 529 DTVSSLALSKDGTFLYSVSWDRTIKVWR-TKDFACLESVRDAHDDAINAVAVSYDG-YVY 702
Query: 84 SGGCDKQVKMWPLLSGGQPM----TVAMHDAPVKDIAWIPEMSLLVTGSWDRTI 133
+G DK++++W L G + T+ H++ + +A + S+L +G+ DR+I
Sbjct: 703 TGSADKRIRVWKKLEGEKKHSLVDTLEKHNSGINALALSADGSVLYSGACDRSI 864
Score = 33.9 bits (76), Expect = 0.11
Identities = 29/119 (24%), Positives = 48/119 (39%), Gaps = 8/119 (6%)
Frame = +1
Query: 64 ISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVA-MHDAPVKDIAWIPEMS 122
+ H V A DGT ++S D+ +K+W +V HD + +A +
Sbjct: 517 VHHVDTVSSLALSKDGTFLYSVSWDRTIKVWRTKDFACLESVRDAHDDAINAVA-VSYDG 693
Query: 123 LLVTGSWDRTIKYWDTRQPNPVHT-------QQLPERCYAMSVKHPLMVVGTADRNILV 174
+ TGS D+ I+ W + H+ A+S ++ G DR+ILV
Sbjct: 694 YVYTGSADKRIRVWKKLEGEKKHSLVDTLEKHNSGINALALSADGSVLYSGACDRSILV 870
>TC229333 similar to UP|Q6H976 (Q6H976) STYLOSA protein, partial (32%)
Length = 982
Score = 54.7 bits (130), Expect = 6e-08
Identities = 28/78 (35%), Positives = 41/78 (51%), Gaps = 1/78 (1%)
Frame = +3
Query: 70 VLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSW 129
V C + DG + SGG DK+V +W S Q T+ H + + D+ + P M L T S+
Sbjct: 669 VACCHFSSDGKLLASGGHDKRVVLWYTDSLKQKATLEEHSSLITDVRFSPSMPRLATSSF 848
Query: 130 DRTIKYWDTRQPN-PVHT 146
D+T+ WD P +HT
Sbjct: 849 DKTVXVWDVDNPGYSLHT 902
>TC229098 similar to PIR|T46032|T46032 WD-40 repeat regulatory protein tup1
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (59%)
Length = 580
Score = 54.3 bits (129), Expect = 8e-08
Identities = 29/108 (26%), Positives = 53/108 (48%), Gaps = 1/108 (0%)
Frame = +3
Query: 31 FSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQ 90
F+P++N++V+ S+D VR W+V K +H PV + DG+ + S D
Sbjct: 78 FNPQSNIIVSGSFDETVRVWDVKSGKCL--KVLPAHSDPVTAVDFNRDGSLIVSSSYDGL 251
Query: 91 VKMWPLLSGGQPMTVAMHD-APVKDIAWIPEMSLLVTGSWDRTIKYWD 137
++W +G T+ D PV + + P ++ G+ D T++ W+
Sbjct: 252 CRIWDASTGHCMKTLIDDDNPPVSFVKFSPNAKFILVGTLDNTLRLWN 395
Score = 47.0 bits (110), Expect = 1e-05
Identities = 32/133 (24%), Positives = 55/133 (41%), Gaps = 4/133 (3%)
Frame = +3
Query: 48 RCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAM 107
R W+V + K H V C + + SG D+ V++W + SG +
Sbjct: 3 RLWDVP--TGSLIKTLHGHTNYVFCVNFNPQSNIIVSGSFDETVRVWDVKSGKCLKVLPA 176
Query: 108 HDAPVKDIAWIPEMSLLVTGSWDRTIKYWDTRQPNPVHT----QQLPERCYAMSVKHPLM 163
H PV + + + SL+V+ S+D + WD + + T P S +
Sbjct: 177 HSDPVTAVDFNRDGSLIVSSSYDGLCRIWDASTGHCMKTLIDDDNPPVSFVKFSPNAKFI 356
Query: 164 VVGTADRNILVYN 176
+VGT D + ++N
Sbjct: 357 LVGTLDNTLRLWN 395
Score = 35.8 bits (81), Expect = 0.029
Identities = 22/76 (28%), Positives = 32/76 (41%), Gaps = 4/76 (5%)
Frame = +3
Query: 235 HREGNEIYSVNSLNFHPVHHTFATAGSDGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNND 294
H N ++ VN F+P + + D WD S + LK + S P+ FN D
Sbjct: 45 HGHTNYVFCVN---FNPQSNIIVSGSFDETVRVWDVKSGKCLKVLPAHSDPVTAVDFNRD 215
Query: 295 GSIFAYS----VCYDW 306
GS+ S +C W
Sbjct: 216 GSLIVSSSYDGLCRIW 263
>TC207996 UP|Q6T2Z5 (Q6T2Z5) WD-repeat cell cycle regulatory protein, complete
Length = 2059
Score = 53.9 bits (128), Expect = 1e-07
Identities = 43/155 (27%), Positives = 69/155 (43%), Gaps = 7/155 (4%)
Frame = +1
Query: 64 ISHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSL 123
+ H V W D + SGG D Q+ +W S + + H A VK IAW P S
Sbjct: 1099 VGHKSEVCGLKWSCDDRELASGGNDNQLLVWNQHSQQPVLRLTEHTAAVKAIAWSPHQSS 1278
Query: 124 LVT---GSWDRTIKYWDT---RQPNPVHT-QQLPERCYAMSVKHPLMVVGTADRNILVYN 176
L+ G+ DR I++W+T Q N V T Q+ ++ +V + G + I+V+
Sbjct: 1279 LLVSGGGTADRCIRFWNTTNGHQLNCVDTGSQVCNLAWSKNVNELVSTHGYSQNQIMVW- 1455
Query: 177 LQNPQVEFKRIISPLKYQTRCLAAFPDQQGFLVGS 211
+ P + ++ + LA PD Q + G+
Sbjct: 1456 -KYPSLTKVATLTGHSMRVLYLAMSPDGQTIVTGA 1557
Score = 37.4 bits (85), Expect = 0.010
Identities = 28/126 (22%), Positives = 51/126 (40%), Gaps = 6/126 (4%)
Frame = +1
Query: 25 SVSSLCFSPKANLLVATSWDNQVRCWEVARNVATAPKASISHDQPVLCSAWKDDGTTVFS 84
+V ++ +SP + L+ + RC + V AW + + S
Sbjct: 1240 AVKAIAWSPHQSSLLVSGGGTADRCIRFWNTTNGHQLNCVDTGSQVCNLAWSKNVNELVS 1419
Query: 85 --GGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWDT---- 138
G Q+ +W S + T+ H V +A P+ +VTG+ D T+++W+
Sbjct: 1420 THGYSQNQIMVWKYPSLTKVATLTGHSMRVLYLAMSPDGQTIVTGAGDETLRFWNVFPSM 1599
Query: 139 RQPNPV 144
+ P PV
Sbjct: 1600 KAPAPV 1617
>BE661627 similar to GP|2443881|gb contains beta-transducin motif
{Arabidopsis thaliana}, partial (13%)
Length = 735
Score = 52.8 bits (125), Expect = 2e-07
Identities = 26/60 (43%), Positives = 34/60 (56%)
Frame = +2
Query: 78 DGTTVFSGGCDKQVKMWPLLSGGQPMTVAMHDAPVKDIAWIPEMSLLVTGSWDRTIKYWD 137
DG V SGG D VK+W L +G H+ ++ I + P LL TGS DRT+K+WD
Sbjct: 11 DGRWVVSGGFDNVVKVWDLTAGKLLHDFKFHEGHIRSIDFHPLEFLLATGSADRTVKFWD 190
Score = 35.0 bits (79), Expect = 0.050
Identities = 24/94 (25%), Positives = 43/94 (45%)
Frame = +2
Query: 202 PDQQGFLVGSIEGRVGVHHLDDSQQGKNFTFKCHREGNEIYSVNSLNFHPVHHTFATAGS 261
PD + + G + V V L + +F F EG+ + S++FHP+ AT +
Sbjct: 8 PDGRWVVSGGFDNVVKVWDLTAGKLLHDFKF---HEGH----IRSIDFHPLEFLLATGSA 166
Query: 262 DGAFNFWDKDSKQRLKAMLRCSQPIPCSAFNNDG 295
D FWD ++ + + + + + AF+ DG
Sbjct: 167 DRTVKFWDLETFELIGSARPEATGVRSIAFHPDG 268
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.132 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,981,027
Number of Sequences: 63676
Number of extensions: 301913
Number of successful extensions: 1705
Number of sequences better than 10.0: 130
Number of HSP's better than 10.0 without gapping: 1506
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1648
length of query: 344
length of database: 12,639,632
effective HSP length: 98
effective length of query: 246
effective length of database: 6,399,384
effective search space: 1574248464
effective search space used: 1574248464
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0314.9


