

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
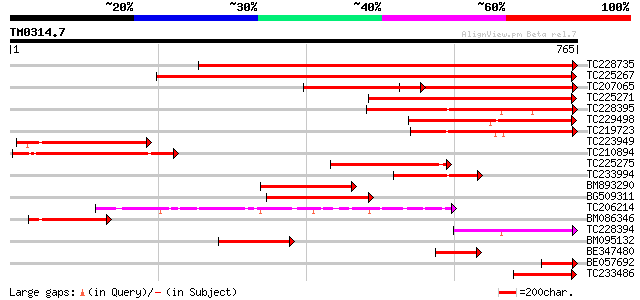
Query= TM0314.7
(765 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC228735 similar to UP|Q76MS5 (Q76MS5) LEXYL1 protein, partial (... 898 0.0
TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucos... 671 0.0
TC207065 beta-glucosidase 447 e-125
TC225271 similar to UP|Q6RXY3 (Q6RXY3) Beta xylosidase (Fragment... 293 3e-79
TC228395 weakly similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_3... 230 2e-60
TC229498 similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, parti... 222 5e-58
TC219723 weakly similar to UP|Q9LXA8 (Q9LXA8) Beta-xylosidase-li... 201 1e-51
TC223949 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, part... 201 1e-51
TC210894 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (25%) 186 3e-47
TC225275 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylos... 166 3e-41
TC233994 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partia... 164 2e-40
BM893290 similar to GP|19879972|gb beta-D-xylosidase {Prunus per... 153 2e-37
BG509311 similar to GP|19879972|gb beta-D-xylosidase {Prunus per... 145 6e-35
TC206214 beta-D-glucan exohydrolase [Glycine max] 137 2e-32
BM086346 similar to GP|18086336|gb| At1g78060/F28K19_32 {Arabido... 119 7e-27
TC228394 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, part... 112 8e-25
BM095132 similar to GP|18025342|gb| beta-D-xylosidase {Hordeum v... 107 2e-23
BE347480 similar to GP|19879972|gb| beta-D-xylosidase {Prunus pe... 92 1e-18
BE057692 similar to GP|10178060|dbj beta-xylosidase {Arabidopsis... 88 2e-17
TC233486 similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, parti... 83 5e-16
>TC228735 similar to UP|Q76MS5 (Q76MS5) LEXYL1 protein, partial (65%)
Length = 1599
Score = 898 bits (2320), Expect = 0.0
Identities = 437/511 (85%), Positives = 473/511 (92%)
Frame = +1
Query: 255 VASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEA 314
VASVMCSYN+VNGKPTCADPDLLKGV+RG+WKLNGYIVSDCDSVEVL+KDQHYTKTPEEA
Sbjct: 1 VASVMCSYNKVNGKPTCADPDLLKGVVRGEWKLNGYIVSDCDSVEVLYKDQHYTKTPEEA 180
Query: 315 AAKSILAGLDLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLP 374
AA SILAGLDL+CG +LG+YTEGAVKQGL+DEASINNA++NNFATLMRLGFFDG+P P
Sbjct: 181 AAISILAGLDLNCGRFLGQYTEGAVKQGLIDEASINNAVTNNFATLMRLGFFDGDPRKQP 360
Query: 375 YGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMI 434
YG LGPKDVCT ENQELAREAARQGIVLLKNSP SLPL+AK+IKSLAVIGPNANATRVMI
Sbjct: 361 YGNLGPKDVCTQENQELAREAARQGIVLLKNSPASLPLNAKAIKSLAVIGPNANATRVMI 540
Query: 435 GNYEGIPCKYISPLQGLTALVPTSYVPGCPDVHCASAQLDDATKIAASSDATVIVVGASL 494
GNYEGIPCKYISPLQGLTA PTSY GC DV C + LDDA KIAAS+DATV+VVGASL
Sbjct: 541 GNYEGIPCKYISPLQGLTAFAPTSYAAGCLDVRCPNPVLDDAKKIAASADATVMVVGASL 720
Query: 495 AIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWV 554
AIEAESLDRVNI+LPGQQQLLVSEVANAS+GPVILVIMSGGGMDVSFAK N+KITSILWV
Sbjct: 721 AIEAESLDRVNILLPGQQQLLVSEVANASKGPVILVIMSGGGMDVSFAKNNNKITSILWV 900
Query: 555 GYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRF 614
GYPGEAGGAAIADVIFGF+NPSGRLPMTWYP+SYV+ VPMTNMNMR DP+TGYPGRTYRF
Sbjct: 901 GYPGEAGGAAIADVIFGFHNPSGRLPMTWYPQSYVDKVPMTNMNMRPDPATGYPGRTYRF 1080
Query: 615 YKGETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSECKSLDVADEHCQNMAFD 674
YKGETVF+FGDG+ YS++ HK+VKAPQLV V LAEDHVCRSSECKS+DV EHCQN+ FD
Sbjct: 1081YKGETVFAFGDGLSYSSIVHKLVKAPQLVSVQLAEDHVCRSSECKSIDVVGEHCQNLVFD 1260
Query: 675 IHLRVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDL 734
IHLR+KN GKMSS+HTV LF TPPAVHNAPQKHLLG EKVHL GKSEA V F VDVCKDL
Sbjct: 1261IHLRIKNKGKMSSAHTVFLFSTPPAVHNAPQKHLLGFEKVHLIGKSEALVSFKVDVCKDL 1440
Query: 735 SVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
S+VDELGNR+V LGQHLLHVG+LK+PLSV I
Sbjct: 1441SIVDELGNRKVALGQHLLHVGDLKHPLSVMI 1533
>TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucosidase,
partial (69%)
Length = 2006
Score = 671 bits (1732), Expect = 0.0
Identities = 325/571 (56%), Positives = 420/571 (72%), Gaps = 4/571 (0%)
Frame = +3
Query: 198 DGDSNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDTFQPPFKSCVIDGNVAS 257
D N LKVAACCKHYTAYD+DNW GV R+ FNA VS+QDL+DT+ PFK+CV++G VAS
Sbjct: 9 DSAGNHLKVAACCKHYTAYDLDNWNGVDRFHFNAKVSKQDLEDTYDVPFKACVLEGQVAS 188
Query: 258 VMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAK 317
VMCSYNQVNGKPTCADPDLL+ IRGQW+LNGYIVSDCDSV V F +QHYTKTPEEAAA+
Sbjct: 189 VMCSYNQVNGKPTCADPDLLRNTIRGQWRLNGYIVSDCDSVGVFFDNQHYTKTPEEAAAE 368
Query: 318 SILAGLDLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGG 377
+I AGLDLDCG +L +T+ A+++GL+ E +N A++N + MRLG FDG PST PYG
Sbjct: 369 AIKAGLDLDCGPFLAIHTDSAIRKGLISENDLNLALANLISVQMRLGMFDGEPSTQPYGN 548
Query: 378 LGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGNY 437
LGP+DVCTS +Q+LA EAAR+ IVLL+N SLPLS ++++ V+GPNA+AT MIGNY
Sbjct: 549 LGPRDVCTSAHQQLALEAARESIVLLQNKGNSLPLSPSRLRTIGVVGPNADATVTMIGNY 728
Query: 438 EGIPCKYISPLQGLTALVPTSYVPGCPDVHCASAQLDDATK-IAASSDATVIVVGASLAI 496
G+ C Y +PLQG+ V T++ GC V C +L A + IA +DA V+V+G +
Sbjct: 729 AGVACGYTTPLQGIARYVKTAHQVGCKGVACRGNELFGAAETIARQADAIVLVMGLDQTV 908
Query: 497 EAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGY 556
EAE+ DRV ++LPG QQ LV+ VA A++GPVIL+IMSGG +D+SFAK + KI++ILWVGY
Sbjct: 909 EAETRDRVGLLLPGLQQELVTRVARAAKGPVILLIMSGGPVDISFAKNDPKISAILWVGY 1088
Query: 557 PGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYK 616
PG+AGG AIADVIFG NP GRLPMTWYP+ Y+ VPMTNM+MR +P+TGYPGRTYRFYK
Sbjct: 1089PGQAGGTAIADVIFGTTNPGGRLPMTWYPQGYLAKVPMTNMDMRPNPTTGYPGRTYRFYK 1268
Query: 617 GETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSS--ECKSLDVADEHC-QNMAF 673
G VF FG G+ YS H + AP+ V VP+ +S K++ V+ +C ++
Sbjct: 1269GPVVFPFGHGLSYSRFSHSLALAPKQVSVPIMSLQALTNSTLSSKAVKVSHANCDDSLEM 1448
Query: 674 DIHLRVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKD 733
+ H+ VKN G M +HT+L+F PP + K L+G K H+ S+ +V+ V VCK
Sbjct: 1449EFHVDVKNEGSMDGTHTLLIFSQPPHGKWSQIKQLVGFHKTHVLAGSKQRVKVGVHVCKH 1628
Query: 734 LSVVDELGNRRVPLGQHLLHVGNLKYPLSVR 764
LSVVD+ G RR+P G+H LH+G++K+ +SV+
Sbjct: 1629LSVVDQFGVRRIPTGEHELHIGDVKHSISVQ 1721
>TC207065 beta-glucosidase
Length = 1328
Score = 447 bits (1149), Expect = e-125
Identities = 212/239 (88%), Positives = 226/239 (93%)
Frame = +2
Query: 527 VILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPE 586
VILVIMSGGGMDVSFAK+NDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYP+
Sbjct: 488 VILVIMSGGGMDVSFAKSNDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPQ 667
Query: 587 SYVENVPMTNMNMRADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVFVP 646
+YV VPMTNMNMRADP+TGYPGRTYRFYKGETVFSFGDGI +S++EHKIVKAPQLV VP
Sbjct: 668 AYVNKVPMTNMNMRADPATGYPGRTYRFYKGETVFSFGDGISFSSIEHKIVKAPQLVSVP 847
Query: 647 LAEDHVCRSSECKSLDVADEHCQNMAFDIHLRVKNMGKMSSSHTVLLFFTPPAVHNAPQK 706
LAEDH CRSSEC SLD+ADEHCQN+AFDIHL VKN GKMS+SH VLLFFTPP VHNAPQK
Sbjct: 848 LAEDHECRSSECMSLDIADEHCQNLAFDIHLGVKNTGKMSTSHVVLLFFTPPDVHNAPQK 1027
Query: 707 HLLGIEKVHLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
HLLG EKVHL GKSEAQVRF VDVCKDLSVVDELGNR+VPLGQHLLHVGNLK+PLS+R+
Sbjct: 1028HLLGFEKVHLPGKSEAQVRFKVDVCKDLSVVDELGNRKVPLGQHLLHVGNLKHPLSLRV 1204
Score = 290 bits (743), Expect = 1e-78
Identities = 144/164 (87%), Positives = 159/164 (96%)
Frame = +1
Query: 397 RQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVP 456
RQGIVLLKNSPGSLPL+AK+IKSLAVIGPNANATRVMIGNYEGIPC YISPLQ LTALVP
Sbjct: 4 RQGIVLLKNSPGSLPLNAKTIKSLAVIGPNANATRVMIGNYEGIPCNYISPLQTLTALVP 183
Query: 457 TSYVPGCPDVHCASAQLDDATKIAASSDATVIVVGASLAIEAESLDRVNIVLPGQQQLLV 516
TSY GCP+V CA+A+LDDAT+IAAS+DATVI+VGASLAIEAESLDR+NI+LPGQQQLLV
Sbjct: 184 TSYAAGCPNVQCANAELDDATQIAASADATVIIVGASLAIEAESLDRINILLPGQQQLLV 363
Query: 517 SEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEA 560
SEVANAS+GPVILVIMSGGGMDVSFAK+NDKITSILW+GYPG++
Sbjct: 364 SEVANASKGPVILVIMSGGGMDVSFAKSNDKITSILWIGYPGDS 495
>TC225271 similar to UP|Q6RXY3 (Q6RXY3) Beta xylosidase (Fragment), partial
(55%)
Length = 952
Score = 293 bits (749), Expect = 3e-79
Identities = 147/283 (51%), Positives = 196/283 (68%), Gaps = 3/283 (1%)
Frame = +3
Query: 485 ATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKA 544
ATV+V+G IEAE+ DRV ++LPG QQ LV+ VA A++GPVILVIMSGG +DVSFAK
Sbjct: 3 ATVLVMGLDQTIEAETRDRVGLLLPGLQQELVTRVARAAKGPVILVIMSGGPVDVSFAKN 182
Query: 545 NDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPS 604
N KI++ILWVGYPG+AGG AIADVIFG NP GRLPMTWYP+ Y+ VPMTNM+MR +P+
Sbjct: 183 NPKISAILWVGYPGQAGGTAIADVIFGATNPGGRLPMTWYPQGYLAKVPMTNMDMRPNPA 362
Query: 605 TGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSS--ECKSLD 662
TGYPGRTYRFYKG VF FG G+ YS + AP+ V V + +S K++
Sbjct: 363 TGYPGRTYRFYKGPVVFPFGHGLSYSRFSQSLALAPKQVSVQILSLQALTNSTLSSKAVK 542
Query: 663 VADEHC-QNMAFDIHLRVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSE 721
V+ +C ++ + H+ VKN G M +HT+L+F PP + K L+ K H+ S+
Sbjct: 543 VSHANCDDSLETEFHVDVKNEGSMDGTHTLLIFSKPPPGKWSQIKQLVTFHKTHVPAGSK 722
Query: 722 AQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVR 764
+++ V CK LSVVD+ G RR+P G+H LH+G+LK+ ++V+
Sbjct: 723 QRLKVNVHSCKHLSVVDQFGVRRIPTGEHELHIGDLKHSINVQ 851
>TC228395 weakly similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, partial
(22%)
Length = 1011
Score = 230 bits (587), Expect = 2e-60
Identities = 122/294 (41%), Positives = 181/294 (61%), Gaps = 10/294 (3%)
Frame = +3
Query: 482 SSDATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSF 541
SS V+V+G + E ES DR + LPG+Q+ L+ VA AS+ PV+LV++ GG +D++
Sbjct: 6 SSRPVVLVMGLDQSQERESHDREYLGLPGKQEELIKSVARASKRPVVLVLLCGGPVDITS 185
Query: 542 AKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRA 601
AK +DK+ ILW GYPGE GG A+A V+FG +NP G+LP+TWYP+ +++ VPMT+M MRA
Sbjct: 186 AKFDDKVGGILWAGYPGELGGVALAQVVFGDHNPGGKLPITWYPKDFIK-VPMTDMRMRA 362
Query: 602 DPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVK-APQLVFVPLAEDH-VCRSSECK 659
DP++GYPGRTYRFY G V+ FG G+ Y+ +K++ + + + + H + ++SE
Sbjct: 363 DPASGYPGRTYRFYTGPKVYEFGYGLSYTKYSYKLLSLSHSTLHINQSSTHLMTQNSETI 542
Query: 660 SL----DVADEHCQNMAFDIHLRVKNMGKMSSSHTVLLFFTPPAVHNA----PQKHLLGI 711
++A+E CQ M I L V N G ++ H VLLF V N P K L+G
Sbjct: 543 RYKLVSELAEETCQTMLLSIALGVTNRGNLAGKHPVLLFVRQGKVRNINNGNPVKQLVGF 722
Query: 712 EKVHLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
+ V + QV F + C+ LSV +E G+ + G +L VG+ +YP+ V +
Sbjct: 723 QSVKVNAGETVQVGFELSPCEHLSVANEAGSMVIEEGSYLFIVGDQEYPIEVTV 884
>TC229498 similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, partial (24%)
Length = 792
Score = 222 bits (565), Expect = 5e-58
Identities = 107/232 (46%), Positives = 153/232 (65%), Gaps = 5/232 (2%)
Frame = +1
Query: 538 DVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNM 597
D++FAK N +I ILW GYPG+AGGAAIAD++FG NP G+LP+TWYPE Y+ +PMTNM
Sbjct: 1 DITFAKNNPRIVGILWAGYPGQAGGAAIADILFGTANPGGKLPVTWYPEEYLTKLPMTNM 180
Query: 598 NMRADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVFVPL---AEDHVCR 654
MRA S GYPGRTYRFY G V+ FG G+ Y++ H + AP +V VPL +V
Sbjct: 181 AMRATKSAGYPGRTYRFYNGPVVYPFGHGLTYTHFVHTLASAPTVVSVPLNGHRRANVTN 360
Query: 655 SSECKSLDVADEHCQNMAFDIHLRVKNMGKMSSSHTVLLFFTPPA--VHNAPQKHLLGIE 712
S +++ V C ++ + + +KN+G +HT+L+F PPA H A +K L+ E
Sbjct: 361 ISN-RAIRVTHARCDKLSISLEVDIKNVGSRDGTHTLLVFSAPPAGFGHWALEKQLVAFE 537
Query: 713 KVHLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVR 764
K+H+ K +V + VCK LSVVD+ G RR+PLG+H ++G++K+ +S++
Sbjct: 538 KIHVPAKGLQRVGVNIHVCKLLSVVDKSGIRRIPLGEHSFNIGDVKHSVSLQ 693
>TC219723 weakly similar to UP|Q9LXA8 (Q9LXA8) Beta-xylosidase-like protein
(AT5g10560/F12B17_90), partial (19%)
Length = 1013
Score = 201 bits (511), Expect = 1e-51
Identities = 98/237 (41%), Positives = 144/237 (60%), Gaps = 12/237 (5%)
Frame = +1
Query: 541 FAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMR 600
FA+ N +I SI+W+GYPGEAGG A+A++IFG +NP+GRLPMTWYPE++ NVPM M+MR
Sbjct: 1 FAEKNPQIASIIWLGYPGEAGGKALAEIIFGEFNPAGRLPMTWYPEAFT-NVPMNEMSMR 177
Query: 601 ADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCR------ 654
ADPS GYPGRTYRFY G V+ FG G+ +S+ + + AP + + R
Sbjct: 178 ADPSRGYPGRTYRFYTGGRVYGFGHGLSFSDFSYNFLSAPSKISLSRTIKDGSRKRLLYQ 357
Query: 655 -SSECKSLDVAD----EHCQNMAFDIHLRVKNMGKMSSSHTVLLFFT-PPAVHNAPQKHL 708
+E +D ++C ++F +H+ V N+G + SH V+LF P V +P+ L
Sbjct: 358 VENEVYGVDYVPVNQLQNCNKLSFSVHISVMNLGGLDGSHVVMLFSKGPKVVDGSPETQL 537
Query: 709 LGIEKVHLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
+G ++H + +V C+ LS D+ G R +PLG H L VG+L++ +S+ I
Sbjct: 538 VGFSRLHTISSKPTETSILVHPCEHLSFADKQGKRILPLGPHTLSVGDLEHVVSIEI 708
>TC223949 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, partial (20%)
Length = 638
Score = 201 bits (510), Expect = 1e-51
Identities = 97/186 (52%), Positives = 127/186 (68%), Gaps = 4/186 (2%)
Frame = +3
Query: 10 SVLVCFSFFCATV---CGQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLVKRLTLQ 66
S + SF T+ T PP++CD + N+ + + FC+ LP+ R DLV RLTL
Sbjct: 12 SAAIFISFLLLTLHHHAESTQPPYSCDSSSNSPY--YPFCNTRLPITKRAQDLVSRLTLD 185
Query: 67 EKIGNLGDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASF 126
EK+ L ++A + RLGIP Y+WWSEALHGV++ G G RF+ + ATSFP ILTAASF
Sbjct: 186 EKLAQLVNTAPAIPRLGIPSYQWWSEALHGVADAGFGIRFNGTIKSATSFPQVILTAASF 365
Query: 127 NSTLFQAIGKVVSTEARAMYNVGLA-GLTYWSPNINIFRDPRWGRGQETPGEDPLLSSKY 185
+ L+ I K + EARA+YN G A G+T+W+PNIN+FRDPRWGRGQET GEDPL+++KY
Sbjct: 366 DPNLWYQISKTIGKEARAVYNAGQATGMTFWAPNINVFRDPRWGRGQETAGEDPLMNAKY 545
Query: 186 AAGYVK 191
YVK
Sbjct: 546 GVAYVK 563
>TC210894 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (25%)
Length = 738
Score = 186 bits (472), Expect = 3e-47
Identities = 109/226 (48%), Positives = 137/226 (60%), Gaps = 2/226 (0%)
Frame = +3
Query: 4 LLQNRVSVLVCFSFFCATVCGQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLVKRL 63
LL + + V +FF G+ PFACD +N GF FC+ +P+ RV DL+ RL
Sbjct: 66 LLALGLVLCVTLTFFPRVTEGRV--PFACD-PRNGLTRGFKFCNTHVPIHVRVQDLIARL 236
Query: 64 TLQEKIGNLGDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTA 123
TL EKI + ++A+ V RLGI YEWWSEALHGVSN+GPGT+F PGAT FP I TA
Sbjct: 237 TLPEKIRLVVNNAIAVPRLGIQGYEWWSEALHGVSNVGPGTKFGGAFPGATMFPQVISTA 416
Query: 124 ASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQETPGEDPLLSS 183
ASFN +L+Q IG+VVS EARAMYN G AGLTYWSPN+NIFRDP G P +
Sbjct: 417 ASFNQSLWQEIGRVVSDEARAMYNGGQAGLTYWSPNVNIFRDPPVGPWPRDSRRGPDIGR 596
Query: 184 KYAAGYVKGLQQTDDGDSNKL--KVAACCKHYTAYDVDNWKGVQRY 227
K + Q+T ++ ++ A CKHY A + GV R+
Sbjct: 597 KIRR---QLCQRTPRRWCPEIASRLLAWCKHYMAMILIIGNGVDRF 725
>TC225275 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosidase
{Prunus persica;} , partial (32%)
Length = 933
Score = 166 bits (421), Expect = 3e-41
Identities = 91/164 (55%), Positives = 114/164 (69%), Gaps = 1/164 (0%)
Frame = +3
Query: 433 MIGNYEGIPCKYISPLQGLTALVPTSYVPGCPDVHCASAQLDDATKIAASS-DATVIVVG 491
MIGNY G+ C Y +PLQG+ V T++ GC V C +L A +I A DATV+V+G
Sbjct: 12 MIGNYAGVACGYTTPLQGIARYVKTAHQVGCRGVACRGNELFGAAEIIARQVDATVLVMG 191
Query: 492 ASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSI 551
IEAE+ DRV ++LPG QQ LV+ VA A++GPVILVIMSGG +DVSFAK N KI++I
Sbjct: 192 LDQTIEAETRDRVGLLLPGLQQELVTRVARAAKGPVILVIMSGGPVDVSFAKNNPKISAI 371
Query: 552 LWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMT 595
LWVGYPG+AGG AIADVIFG NP G+L + Y+E + T
Sbjct: 372 LWVGYPGQAGGTAIADVIFGATNP-GKL----FSHHYLEFISYT 488
>TC233994 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partial (15%)
Length = 462
Score = 164 bits (414), Expect = 2e-40
Identities = 75/119 (63%), Positives = 93/119 (78%)
Frame = +2
Query: 519 VANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGR 578
VA AS+ PVILV++SGG +D++ AK N KI ILW GYPGE GG A+A +IFG +NP GR
Sbjct: 2 VAEASKKPVILVLLSGGPLDITSAKYNHKIGGILWAGYPGELGGIALAQIIFGDHNPGGR 181
Query: 579 LPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIV 637
LP TWYP+ Y++ VPMT+M MRADPSTGYPGRTYRFYKG V+ FG G+ YS +++V
Sbjct: 182 LPTTWYPKDYIK-VPMTDMRMRADPSTGYPGRTYRFYKGPKVYEFGYGLSYSKYSYELV 355
>BM893290 similar to GP|19879972|gb beta-D-xylosidase {Prunus persica},
partial (27%)
Length = 403
Score = 153 bits (387), Expect = 2e-37
Identities = 75/130 (57%), Positives = 90/130 (68%)
Frame = +1
Query: 339 VKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQ 398
VK+GL+ EA +N A+ N MRLG +DG PS+ PY LGP+DVCT +QELA EAARQ
Sbjct: 1 VKKGLISEADVNGALLNTLTVQMRLGMYDGEPSSHPYNNLGPRDVCTQSHQELALEAARQ 180
Query: 399 GIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTS 458
GIVLLKN SLPLS + +++AVIGPN+N T MIGNY GI C Y SPLQG+ T
Sbjct: 181 GIVLLKNKGPSLPLSTRRGRTVAVIGPNSNVTFTMIGNYAGIACGYTSPLQGIGTYTKTI 360
Query: 459 YVPGCPDVHC 468
Y GC +V C
Sbjct: 361 YEHGCANVAC 390
>BG509311 similar to GP|19879972|gb beta-D-xylosidase {Prunus persica},
partial (31%)
Length = 443
Score = 145 bits (366), Expect = 6e-35
Identities = 76/146 (52%), Positives = 97/146 (66%), Gaps = 1/146 (0%)
Frame = +1
Query: 347 ASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNS 406
A +N A+ N MRLG FDG P+ PYG LGPKDVC +QELA EAARQGIVLLKN+
Sbjct: 1 ADVNGALVNTLTVQMRLGMFDGEPTAHPYGHLGPKDVCKPAHQELALEAARQGIVLLKNT 180
Query: 407 PGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPGCPDV 466
LPLS++ +++AVIGPN+ AT MIGNY G+ C Y +PLQG+ T + GC +V
Sbjct: 181 GPVLPLSSQLHRTVAVIGPNSKATITMIGNYAGVACGYTNPLQGIGRYARTVHQLGCQNV 360
Query: 467 HCASAQL-DDATKIAASSDATVIVVG 491
C + +L A A +DATV+V+G
Sbjct: 361 ACKNDKLFGPAINAARQADATVLVMG 438
>TC206214 beta-D-glucan exohydrolase [Glycine max]
Length = 1647
Score = 137 bits (345), Expect = 2e-32
Identities = 148/521 (28%), Positives = 226/521 (42%), Gaps = 33/521 (6%)
Frame = +1
Query: 116 FPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTY-WSPNINIFRDPRWGRGQET 174
FP + + + L + IG+ + E RA G+ Y ++P I + RDPRWGR E+
Sbjct: 4 FPHNVGLGVTRDPVLIKKIGEATALEVRA------TGIPYVFAPCIAVCRDPRWGRCYES 165
Query: 175 PGEDPLLSSKYAAGYVKGLQQTDDGDS--------NKLKVAACCKHYTAYDVDNWKGVQR 226
EDP + K + GLQ G+S K KVAAC KHY D KG+
Sbjct: 166 YSEDPKI-VKTMTEIIPGLQGDIPGNSIKGVPFVAGKNKVAACAKHYLG-DGGTNKGINE 339
Query: 227 YTFNAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWK 286
N ++S L P + +I G V++VM SY+ NG A+ L+ G ++ +
Sbjct: 340 N--NTLISYNGLLSIHMPAYYDSIIKG-VSTVMISYSSWNGMKMHANKKLITGYLKNKLH 510
Query: 287 LNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE------GAVK 340
G+++SD ++ + H + A S AG+D+ + YTE VK
Sbjct: 511 FKGFVISDWQGIDRITSPPHANYSYSVQAGVS--AGIDMIMVPF--NYTEFIDELTRQVK 678
Query: 341 QGLLDEASINNAISNNFATLMRLGFFDGNPSTLP--YGGLGPKDVCTSENQELAREAARQ 398
++ + I++A++ +G F+ NP P LG K E++E+AREA R+
Sbjct: 679 NNIIPISRIDDAVARILRVKFVMGLFE-NPYADPSLANQLGSK-----EHREIAREAVRK 840
Query: 399 GIVLLKNSPG----SLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTAL 454
+VLLKN LPL KS K L V G +AN G + QGL
Sbjct: 841 SLVLLKNGKSYKKPLLPLPKKSTKIL-VAGSHANNLGYQCGG-------WTITWQGLGGN 996
Query: 455 VPTSYVPGCPDVHCASAQLDDATKIAASS------------DATVIVVGASLAIEAESLD 502
TS G + +D AT++ + D ++VVG E D
Sbjct: 997 DLTS---GTTILDAVKQTVDPATEVVFNENPDKNFVKSYKFDYAIVVVGEHTYAETFG-D 1164
Query: 503 RVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGG 562
+N+ + ++ V A R V+LV G V KI +++ PG G
Sbjct: 1165SLNLTMADPGPSTITNVCGAIRCVVVLVT----GRPVVIKPYLPKIDALVAAWLPG-TEG 1329
Query: 563 AAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADP 603
+ADV++G Y +G+L TW+ V+ +PM + DP
Sbjct: 1330QGVADVLYGDYEFTGKLARTWF--KTVDQLPMNVGDKHYDP 1446
>BM086346 similar to GP|18086336|gb| At1g78060/F28K19_32 {Arabidopsis
thaliana}, partial (11%)
Length = 427
Score = 119 bits (297), Expect = 7e-27
Identities = 57/112 (50%), Positives = 76/112 (66%)
Frame = -3
Query: 26 TSPPFACDVTKNASFAGFGFCDKSLPVADRVADLVKRLTLQEKIGNLGDSAVDVGRLGIP 85
T PP++CD + N+ + + FC+ LP+ R DLV RLTL EK+ L ++A + RLGIP
Sbjct: 419 TQPPYSCDSSSNSPY--YPFCNTRLPITKRAQDLVSRLTLDEKLAQLVNTAPAIPRLGIP 246
Query: 86 RYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASFNSTLFQAIGKV 137
Y+WWSEALHGV++ G G RF+ + ATSFP ILTAASF+ L+ I KV
Sbjct: 245 SYQWWSEALHGVADAGFGIRFNGTIKSATSFPQVILTAASFDPNLWYQISKV 90
>TC228394 similar to UP|Q8VZG5 (Q8VZG5) At1g78060/F28K19_32, partial (14%)
Length = 659
Score = 112 bits (279), Expect = 8e-25
Identities = 65/175 (37%), Positives = 97/175 (55%), Gaps = 8/175 (4%)
Frame = +2
Query: 599 MRADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVK-APQLVFVPLAEDHV-CRSS 656
MRADP++GYPGRTYRFY G V+ FG G+ Y+ +K++ + + + + H+ ++S
Sbjct: 2 MRADPASGYPGRTYRFYTGPKVYEFGYGLSYTKYSYKLLSLSHNTLHINQSSTHLTTQNS 181
Query: 657 ECKSL----DVADEHCQNMAFDIHLRVKNMGKMSSSHTVLLFFTPPAVHN--APQKHLLG 710
E ++A+E CQ M I L V N G M+ H VLLF V N P K L+G
Sbjct: 182 ETIRYKLVSELAEETCQTMLLSIALGVTNHGNMAGKHPVLLFVRQGKVRNNGNPVKQLVG 361
Query: 711 IEKVHLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
+ V L QV F + C+ LSV +E G+ + G +LL VG+ +YP+ + +
Sbjct: 362 FQSVKLNAGETVQVGFELSPCEHLSVANEAGSMVIEEGSYLLLVGDQEYPIEITV 526
>BM095132 similar to GP|18025342|gb| beta-D-xylosidase {Hordeum vulgare},
partial (2%)
Length = 428
Score = 107 bits (268), Expect = 2e-23
Identities = 48/103 (46%), Positives = 69/103 (66%)
Frame = +1
Query: 282 RGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTEGAVKQ 341
R QWK +GYI SDC +V ++ + Q Y KT E+A A AG+D++CG Y+ K+ + AV Q
Sbjct: 13 RQQWKFDGYITSDCGAVSIIHEKQGYAKTAEDAIADVFRAGMDVECGDYITKHAKSAVFQ 192
Query: 342 GLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVC 384
L + I+ A+ N F+ +RLG FDGNP+ LP+G +GP +VC
Sbjct: 193 KKLPISQIDRALQNLFSIRIRLGLFDGNPTKLPFGTIGPNEVC 321
>BE347480 similar to GP|19879972|gb| beta-D-xylosidase {Prunus persica},
partial (17%)
Length = 473
Score = 91.7 bits (226), Expect = 1e-18
Identities = 37/62 (59%), Positives = 47/62 (75%)
Frame = +3
Query: 575 PSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEH 634
P G+LPMTWYP+ Y++N+PMTNM MRA S GYPGRTYRFY G V+ FG G+ Y++ H
Sbjct: 282 PGGKLPMTWYPQGYIKNLPMTNMAMRASRSKGYPGRTYRFYNGPVVYPFGYGLSYTHFVH 461
Query: 635 KI 636
+
Sbjct: 462 TL 467
Score = 35.0 bits (79), Expect = 0.12
Identities = 14/19 (73%), Positives = 17/19 (88%)
Frame = +2
Query: 557 PGEAGGAAIADVIFGFYNP 575
PG+AGGAAIAD++FG NP
Sbjct: 2 PGQAGGAAIADILFGTSNP 58
>BE057692 similar to GP|10178060|dbj beta-xylosidase {Arabidopsis thaliana},
partial (5%)
Length = 225
Score = 87.8 bits (216), Expect = 2e-17
Identities = 41/48 (85%), Positives = 45/48 (93%)
Frame = +2
Query: 718 GKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
GKSEAQVRF VD+CKDLSVVDELGNR+VPLGQHLLHVGNLK+ LSV +
Sbjct: 5 GKSEAQVRFKVDICKDLSVVDELGNRKVPLGQHLLHVGNLKHQLSVSV 148
>TC233486 similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, partial (7%)
Length = 677
Score = 82.8 bits (203), Expect = 5e-16
Identities = 39/87 (44%), Positives = 59/87 (66%), Gaps = 2/87 (2%)
Frame = +2
Query: 680 KNMGKMSSSHTVLLFFTPPAV--HNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSVV 737
KN+G +HT+L+F PPA H AP K L+ +K+H+ K++ +V + VCK LSVV
Sbjct: 2 KNVGSKDGTHTLLVFSAPPAGNGHWAPHKQLVAFQKLHIPSKAQQRVNVNIHVCKLLSVV 181
Query: 738 DELGNRRVPLGQHLLHVGNLKYPLSVR 764
D G RRVP+G H LH+G++K+ +S++
Sbjct: 182 DRSGTRRVPMGLHSLHIGDVKHYVSLQ 262
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,588,244
Number of Sequences: 63676
Number of extensions: 460323
Number of successful extensions: 2130
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 2097
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2107
length of query: 765
length of database: 12,639,632
effective HSP length: 105
effective length of query: 660
effective length of database: 5,953,652
effective search space: 3929410320
effective search space used: 3929410320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0314.7


