

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0310b.14
(587 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
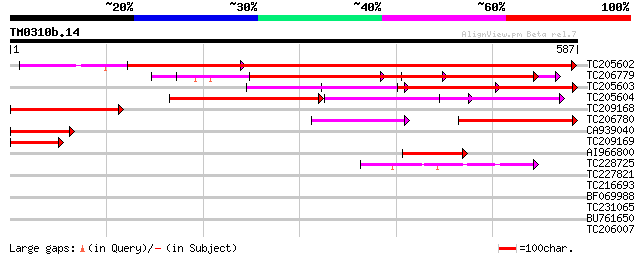
Score E
Sequences producing significant alignments: (bits) Value
TC205602 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protei... 821 0.0
TC206779 homologue to UP|Q9ZRU5 (Q9ZRU5) Protein phosphatase (Fr... 566 e-161
TC205603 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protei... 315 5e-86
TC205604 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protei... 291 7e-79
TC209168 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protei... 219 3e-57
TC206780 similar to UP|2AAA_PEA (P36875) Protein phosphatase PP2... 207 8e-54
CA939040 homologue to PIR|S51808|S518 phosphoprotein phosphatase... 123 2e-28
TC209169 homologue to UP|Q38950 (Q38950) Protein phosphatase 2A ... 99 4e-21
AI966800 similar to GP|11094365|gb| Ser/Thr specific protein pho... 97 3e-20
TC228725 homologue to UP|Q9FMF9 (Q9FMF9) Nuclear protein-like, p... 43 3e-04
TC227821 weakly similar to UP|Q7PY15 (Q7PY15) EbiP8519 (Fragment... 35 0.12
TC216693 30 2.3
BF069988 29 6.6
TC231065 29 6.6
BU761650 29 6.6
TC206007 similar to GB|AAO64828.1|29028898|BT005893 At3g56480 {A... 29 6.6
>TC205602 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protein
phosphatase 2A A regulatory subunit alpha isoform,
partial (79%)
Length = 1680
Score = 821 bits (2121), Expect = 0.0
Identities = 411/464 (88%), Positives = 446/464 (95%)
Frame = +1
Query: 123 VEHFIPLVKRLAAGEWFTARVSSCGLFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRS 182
VE++IPLVKRLAAGEWFTARVS+CGLFHIAYPSAPE K ELR++Y QLCQDDMPMVRRS
Sbjct: 1 VEYYIPLVKRLAAGEWFTARVSACGLFHIAYPSAPETSKTELRSIYSQLCQDDMPMVRRS 180
Query: 183 AATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHI 242
AA+NLGKFAATVE AHLK+D+MS+FDDLTQDDQDSVRLLAVEGCAALGKLLEPQ+CVAHI
Sbjct: 181 AASNLGKFAATVEYAHLKADVMSIFDDLTQDDQDSVRLLAVEGCAALGKLLEPQDCVAHI 360
Query: 243 LPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAG 302
LPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAG
Sbjct: 361 LPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAG 540
Query: 303 KVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPI 362
KVTKF RIL+P+L+I+HIL CVKELS+DSSQHVRSALASVIMGMAPVLGK+ATIEQLLPI
Sbjct: 541 KVTKFCRILNPDLSIQHILSCVKELSSDSSQHVRSALASVIMGMAPVLGKEATIEQLLPI 720
Query: 363 FLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIP 422
FLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIP
Sbjct: 721 FLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIP 900
Query: 423 LLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLD 482
LLASQLGV FFDDKLGALCMQWL+DKV+SIR+AAANN+KRLAEEFGP+WAMQHIIPQVL+
Sbjct: 901 LLASQLGVSFFDDKLGALCMQWLQDKVHSIREAAANNLKRLAEEFGPEWAMQHIIPQVLE 1080
Query: 483 MINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLI 542
M N+PHYLYRMTIL+AISLLAPV+G EIT S LLP+V+ ASKDRVPNIKFNVAKVL+S+
Sbjct: 1081MNNNPHYLYRMTILRAISLLAPVMGPEITCSNLLPVVLAASKDRVPNIKFNVAKVLESIF 1260
Query: 543 PIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMS 586
PIV++SVVE T+RPCLVELSEDPDVDVRFF++QALQ+ D V MS
Sbjct: 1261PIVDQSVVEKTIRPCLVELSEDPDVDVRFFSNQALQAIDHVMMS 1392
Score = 46.2 bits (108), Expect = 4e-05
Identities = 48/240 (20%), Positives = 102/240 (42%), Gaps = 6/240 (2%)
Frame = +1
Query: 11 IAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDD-EVLLAMAE 69
+ + + LK+E +RLN I +L + + +G + + L+P + E +D V LA+ E
Sbjct: 712 LPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIE 891
Query: 70 ELGVFIPYVGGVEHANVLLPPLETLCTV----EETCVRDKSVESLCRIGAQMREQDLVEH 125
+IP + + L LC + +R+ + +L R+ + + ++H
Sbjct: 892 ----YIPLLASQLGVSFFDDKLGALCMQWLQDKVHSIREAAANNLKRLAEEFGPEWAMQH 1059
Query: 126 FIPLVKRLAAGEWFTARVSSCGLFHIAYP-SAPEAQKAELRAMYGQLCQDDMPMVRRSAA 184
IP V + + R++ + P PE + L + +D +P ++ + A
Sbjct: 1060IIPQVLEMNNNPHYLYRMTILRAISLLAPVMGPEITCSNLLPVVLAASKDRVPNIKFNVA 1239
Query: 185 TNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLLEPQECVAHILP 244
L V+ + ++ I +L++D VR + + A+ ++ C A ++P
Sbjct: 1240KVLESIFPIVDQSVVEKTIRPCLVELSEDPDVDVRFFSNQALQAIDHVM--MSC*AEMVP 1413
>TC206779 homologue to UP|Q9ZRU5 (Q9ZRU5) Protein phosphatase (Fragment),
partial (55%)
Length = 897
Score = 566 bits (1458), Expect = e-161
Identities = 286/299 (95%), Positives = 296/299 (98%)
Frame = +1
Query: 249 FSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFS 308
FSQDKSWRVRYMVANQLYELCEAVGP+PTR+ELVPAYVRLLRDNEAEVRIAAAGKVTKFS
Sbjct: 1 FSQDKSWRVRYMVANQLYELCEAVGPDPTRSELVPAYVRLLRDNEAEVRIAAAGKVTKFS 180
Query: 309 RILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLK 368
RIL+P+LAI+HILPCVKELS DSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLK
Sbjct: 181 RILNPDLAIQHILPCVKELSTDSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLK 360
Query: 369 DEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQL 428
DEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQL
Sbjct: 361 DEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQL 540
Query: 429 GVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPH 488
GVGFFDDKLGALCMQWLKDKVYSIRDAAANN+KRLAEEFGPDWAMQHIIPQVLDM+ DPH
Sbjct: 541 GVGFFDDKLGALCMQWLKDKVYSIRDAAANNIKRLAEEFGPDWAMQHIIPQVLDMVTDPH 720
Query: 489 YLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVEE 547
YLYRMTILQAISLLAPVLGSEITSSKLLPLV+NASKDRVPNIKFNV KVLQSLIPIV++
Sbjct: 721 YLYRMTILQAISLLAPVLGSEITSSKLLPLVINASKDRVPNIKFNVXKVLQSLIPIVDQ 897
Score = 119 bits (297), Expect = 5e-27
Identities = 74/281 (26%), Positives = 139/281 (49%)
Frame = +1
Query: 173 QDDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKL 232
QD VR A L + V +S+++ + L +D++ VR+ A ++
Sbjct: 7 QDKSWRVRYMVANQLYELCEAVGPDPTRSELVPAYVRLLRDNEAEVRIAAAGKVTKFSRI 186
Query: 233 LEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDN 292
L P + HILP + S D S VR +A+ + + +G + T +L+P ++ LL+D
Sbjct: 187 LNPDLAIQHILPCVKELSTDSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDE 366
Query: 293 EAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGK 352
+VR+ K+ + ++++ +L + +LP + EL+ D VR A+ I +A LG
Sbjct: 367 FPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGV 546
Query: 353 DATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWR 412
++L + + LKD+ +R + + ++ + G D Q ++P ++++ D H+
Sbjct: 547 GFFDDKLGALCMQWLKDKVYSIRDAAANNIKRLAEEFGPDWAMQHIIPQVLDMVTDPHYL 726
Query: 413 VRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIR 453
R+ I++ I LLA LG KL L + KD+V +I+
Sbjct: 727 YRMTILQAISLLAPVLGSEITSSKLLPLVINASKDRVPNIK 849
Score = 89.7 bits (221), Expect = 3e-18
Identities = 65/281 (23%), Positives = 123/281 (43%), Gaps = 39/281 (13%)
Frame = +1
Query: 148 LFHIAYPSAPEAQKAELRAMYGQLCQDDMPMVRRSAATNLGKFA---------------- 191
L+ + P+ ++EL Y +L +D+ VR +AA + KF+
Sbjct: 49 LYELCEAVGPDPTRSELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILNPDLAIQHILPCV 228
Query: 192 ---ATVEAAHLKSDIMSV--------------------FDDLTQDDQDSVRLLAVEGCAA 228
+T + H++S + SV F L +D+ VRL +
Sbjct: 229 KELSTDSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQ 408
Query: 229 LGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRL 288
+ +++ +LP IV ++D+ WRVR + + L +G +L ++
Sbjct: 409 VNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQW 588
Query: 289 LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAP 348
L+D +R AAA + + + P+ A++HI+P V ++ D R + I +AP
Sbjct: 589 LKDKVYSIRDAAANNIKRLAEEFGPDWAMQHIIPQVLDMVTDPHYLYRMTILQAISLLAP 768
Query: 349 VLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVI 389
VLG + T +LLP+ ++ KD P+++ N+ L + ++
Sbjct: 769 VLGSEITSSKLLPLVINASKDRVPNIKFNVXKVLQSLIPIV 891
Score = 89.0 bits (219), Expect = 5e-18
Identities = 49/165 (29%), Positives = 85/165 (50%)
Frame = +1
Query: 406 AEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAE 465
++D+ WRVR + + L +G +L ++ L+D +R AAA V + +
Sbjct: 4 SQDKSWRVRYMVANQLYELCEAVGPDPTRSELVPAYVRLLRDNEAEVRIAAAGKVTKFSR 183
Query: 466 EFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKD 525
PD A+QHI+P V ++ D R + I +APVLG + T +LLP+ ++ KD
Sbjct: 184 ILNPDLAIQHILPCVKELSTDSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKD 363
Query: 526 RVPNIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVR 570
P+++ N+ L + ++ ++ +L P +VEL+ED VR
Sbjct: 364 EFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 498
Score = 34.3 bits (77), Expect = 0.16
Identities = 35/176 (19%), Positives = 73/176 (40%), Gaps = 6/176 (3%)
Frame = +1
Query: 11 IAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDD-EVLLAMAE 69
+ + + LK+E +RLN I +L + + +G + + L+P + E +D V LA+ E
Sbjct: 334 LPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIE 513
Query: 70 ELGVFIPYVGGVEHANVLLPPLETLCTV----EETCVRDKSVESLCRIGAQMREQDLVEH 125
+IP + L LC + +RD + ++ R+ + ++H
Sbjct: 514 ----YIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNIKRLAEEFGPDWAMQH 681
Query: 126 FIPLVKRLAAGEWFTARVSSCGLFHIAYP-SAPEAQKAELRAMYGQLCQDDMPMVR 180
IP V + + R++ + P E ++L + +D +P ++
Sbjct: 682 IIPQVLDMVTDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVINASKDRVPNIK 849
>TC205603 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protein
phosphatase 2A A regulatory subunit alpha isoform,
partial (32%)
Length = 974
Score = 315 bits (806), Expect = 5e-86
Identities = 155/186 (83%), Positives = 175/186 (93%)
Frame = +2
Query: 402 IVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVK 461
IVELAEDRHWRVRLAIIEYIPLLASQLGVGFF DKLGALCMQWL+DKV+SIR+AAANN+K
Sbjct: 2 IVELAEDRHWRVRLAIIEYIPLLASQLGVGFFYDKLGALCMQWLQDKVHSIREAAANNLK 181
Query: 462 RLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVN 521
RLAEEFGP+WAMQHIIPQVL+MI++PHYLYRMTIL AISLLAPV+GSEIT S+LLP+V+
Sbjct: 182 RLAEEFGPEWAMQHIIPQVLEMISNPHYLYRMTILHAISLLAPVMGSEITRSELLPIVIT 361
Query: 522 ASKDRVPNIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSD 581
ASKDRVPNIKFNVAKVL+S+ PIV++SVVE T+RP LVELSEDPDVDVRFF++QAL + D
Sbjct: 362 ASKDRVPNIKFNVAKVLESIFPIVDQSVVEKTIRPSLVELSEDPDVDVRFFSNQALHAMD 541
Query: 582 QVKMSS 587
V MSS
Sbjct: 542 HVMMSS 559
Score = 89.7 bits (221), Expect = 3e-18
Identities = 48/169 (28%), Positives = 91/169 (53%)
Frame = +2
Query: 246 IVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVT 305
IV ++D+ WRVR + + L +G +L ++ L+D +R AAA +
Sbjct: 2 IVELAEDRHWRVRLAIIEYIPLLASQLGVGFFYDKLGALCMQWLQDKVHSIREAAANNLK 181
Query: 306 KFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLS 365
+ + PE A++HI+P V E+ ++ R + I +APV+G + T +LLPI ++
Sbjct: 182 RLAEEFGPEWAMQHIIPQVLEMISNPHYLYRMTILHAISLLAPVMGSEITRSELLPIVIT 361
Query: 366 LLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 414
KD P+++ N+ L+ + ++ ++ +++ P++VEL+ED VR
Sbjct: 362 ASKDRVPNIKFNVAKVLESIFPIVDQSVVEKTIRPSLVELSEDPDVDVR 508
Score = 53.5 bits (127), Expect = 3e-07
Identities = 42/185 (22%), Positives = 85/185 (45%)
Frame = +2
Query: 324 VKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLD 383
+ EL+ D VR A+ I +A LG ++L + + L+D+ +R + L
Sbjct: 2 IVELAEDRHWRVRLAIIEYIPLLASQLGVGFFYDKLGALCMQWLQDKVHSIREAAANNLK 181
Query: 384 QVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQ 443
++ + G + Q ++P ++E+ + H+ R+ I+ I LLA +G +L + +
Sbjct: 182 RLAEEFGPEWAMQHIIPQVLEMISNPHYLYRMTILHAISLLAPVMGSEITRSELLPIVIT 361
Query: 444 WLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLA 503
KD+V +I+ A ++ + + I P ++++ DP R QA+ +
Sbjct: 362 ASKDRVPNIKFNVAKVLESIFPIVDQSVVEKTIRPSLVELSEDPDVDVRFFSNQALHAMD 541
Query: 504 PVLGS 508
V+ S
Sbjct: 542 HVMMS 556
>TC205604 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protein
phosphatase 2A A regulatory subunit alpha isoform,
partial (27%)
Length = 482
Score = 291 bits (744), Expect = 7e-79
Identities = 141/160 (88%), Positives = 154/160 (96%)
Frame = +3
Query: 166 AMYGQLCQDDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEG 225
+MY LCQDDMPMVRRSAA+NLGK+AATVE AHLK+D MS+F+DLT+DDQDSVRLLAVEG
Sbjct: 3 SMYSLLCQDDMPMVRRSAASNLGKYAATVEYAHLKADTMSIFEDLTKDDQDSVRLLAVEG 182
Query: 226 CAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAY 285
CAALGKLLEPQ+C+ HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAY
Sbjct: 183 CAALGKLLEPQDCITHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAY 362
Query: 286 VRLLRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVK 325
VRLLRDNEAEVRIAAAGKVTKF RIL+P+L+I+HILPCVK
Sbjct: 363 VRLLRDNEAEVRIAAAGKVTKFCRILNPDLSIQHILPCVK 482
Score = 55.8 bits (133), Expect = 5e-08
Identities = 41/154 (26%), Positives = 67/154 (42%)
Frame = +3
Query: 327 LSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVN 386
L D VR + AS + A + + IF L KD+ VRL + +
Sbjct: 18 LCQDDMPMVRRSAASNLGKYAATVEYAHLKADTMSIFEDLTKDDQDSVRLLAVEGCAALG 197
Query: 387 QVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLK 446
+++ +LP IV ++D+ WRVR + + L +G +L ++ L+
Sbjct: 198 KLLEPQDCITHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLR 377
Query: 447 DKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQV 480
D +R AAA V + PD ++QHI+P V
Sbjct: 378 DNEAEVRIAAAGKVTKFCRILNPDLSIQHILPCV 479
Score = 42.4 bits (98), Expect = 6e-04
Identities = 30/129 (23%), Positives = 57/129 (43%)
Frame = +3
Query: 446 KDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPV 505
+D + +R +AA+N+ + A + D+ D R+ ++ + L +
Sbjct: 24 QDDMPMVRRSAASNLGKYAATVEYAHLKADTMSIFEDLTKDDQDSVRLLAVEGCAALGKL 203
Query: 506 LGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDP 565
L + + +LP++VN S+D+ +++ VA L L V L P V L D
Sbjct: 204 LEPQDCITHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDN 383
Query: 566 DVDVRFFAS 574
+ +VR A+
Sbjct: 384 EAEVRIAAA 410
>TC209168 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protein
phosphatase 2A A regulatory subunit alpha isoform,
partial (20%)
Length = 574
Score = 219 bits (558), Expect = 3e-57
Identities = 108/118 (91%), Positives = 116/118 (97%)
Frame = +1
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
++M D+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTR+ELIPFLSENNDDD
Sbjct: 220 ISMADEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRRELIPFLSENNDDD 399
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMR 118
DEVLLAMAEELGVFIPYVGGVEHA+VLLPPLETLCTVEETCVRDK+ ESLCRIG+QMR
Sbjct: 400 DEVLLAMAEELGVFIPYVGGVEHASVLLPPLETLCTVEETCVRDKAAESLCRIGSQMR 573
Score = 29.3 bits (64), Expect = 5.1
Identities = 30/116 (25%), Positives = 53/116 (44%), Gaps = 6/116 (5%)
Frame = +1
Query: 357 EQLLPIFLSL--LKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 414
E L PI + + LK++ +RLN I +L + + +G + + L+P + E +D V
Sbjct: 235 EPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRRELIPFLSENNDDDD-EVL 411
Query: 415 LAIIE----YIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEE 466
LA+ E +IP + L LC + +RD AA ++ R+ +
Sbjct: 412 LAMAEELGVFIPYVGGVEHASVLLPPLETLCTV----EETCVRDKAAESLCRIGSQ 567
>TC206780 similar to UP|2AAA_PEA (P36875) Protein phosphatase PP2A regulatory
subunit A (PR65) (Fragment), partial (31%)
Length = 667
Score = 207 bits (528), Expect = 8e-54
Identities = 106/123 (86%), Positives = 114/123 (92%)
Frame = +2
Query: 465 EEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASK 524
EEF P MQHII V M+ DPHYLYRMTILQ+ISLLAPVLGSEI+SSKLLPLV+NASK
Sbjct: 2 EEFXPXXXMQHIIXXVXXMVTDPHYLYRMTILQSISLLAPVLGSEISSSKLLPLVINASK 181
Query: 525 DRVPNIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVK 584
DRVPNIKFNVAKVLQSLIPIV++SVVE+T+RPCLVELSEDPDVDVRFFASQALQSSDQVK
Sbjct: 182 DRVPNIKFNVAKVLQSLIPIVDQSVVESTIRPCLVELSEDPDVDVRFFASQALQSSDQVK 361
Query: 585 MSS 587
MSS
Sbjct: 362 MSS 370
Score = 56.6 bits (135), Expect = 3e-08
Identities = 28/102 (27%), Positives = 53/102 (51%)
Frame = +2
Query: 313 PELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFP 372
P ++HI+ V + D R + I +APVLG + + +LLP+ ++ KD P
Sbjct: 14 PXXXMQHIIXXVXXMVTDPHYLYRMTILQSISLLAPVLGSEISSSKLLPLVINASKDRVP 193
Query: 373 DVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 414
+++ N+ L + ++ ++ ++ P +VEL+ED VR
Sbjct: 194 NIKFNVAKVLQSLIPIVDQSVVESTIRPCLVELSEDPDVDVR 319
Score = 37.0 bits (84), Expect = 0.024
Identities = 22/102 (21%), Positives = 41/102 (39%)
Frame = +2
Query: 235 PQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEA 294
P + HI+ + D + R + + L +G E + ++L+P + +D
Sbjct: 14 PXXXMQHIIXXVXXMVTDPHYLYRMTILQSISLLAPVLGSEISSSKLLPLVINASKDRVP 193
Query: 295 EVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVR 336
++ A + I+ + I PC+ ELS D VR
Sbjct: 194 NIKFNVAKVLQSLIPIVDQSVVESTIRPCLVELSEDPDVDVR 319
>CA939040 homologue to PIR|S51808|S518 phosphoprotein phosphatase 2A 65K
regulatory chain homolog pDF1 - Arabidopsis thaliana,
partial (11%)
Length = 408
Score = 123 bits (309), Expect = 2e-28
Identities = 61/67 (91%), Positives = 66/67 (98%)
Frame = +1
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
M+MVD+PLYPIAVLIDELKN+DIQLRLNSIR+LSTIARALGEERTR+ELIPFL ENNDDD
Sbjct: 208 MSMVDEPLYPIAVLIDELKNDDIQLRLNSIRKLSTIARALGEERTRRELIPFLGENNDDD 387
Query: 61 DEVLLAM 67
DEVLLAM
Sbjct: 388 DEVLLAM 408
>TC209169 homologue to UP|Q38950 (Q38950) Protein phosphatase 2A 65 kDa
regulatory subunit, partial (9%)
Length = 378
Score = 99.4 bits (246), Expect = 4e-21
Identities = 49/55 (89%), Positives = 54/55 (98%)
Frame = +2
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSE 55
++M D+PLYPIAVLIDELKN+DIQLRLNSIRRLSTIARALGEERTR+ELIPFLSE
Sbjct: 212 ISMADEPLYPIAVLIDELKNDDIQLRLNSIRRLSTIARALGEERTRRELIPFLSE 376
>AI966800 similar to GP|11094365|gb| Ser/Thr specific protein phosphatase 2A
A regulatory subunit alpha isoform, partial (11%)
Length = 208
Score = 96.7 bits (239), Expect = 3e-20
Identities = 51/68 (75%), Positives = 55/68 (80%)
Frame = +2
Query: 407 EDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEE 466
+DRH R RLAIIEYIPL AS GV DDKLGALCMQ L+D V+SIR+AAANN KRLAEE
Sbjct: 5 DDRHCRPRLAIIEYIPLSAS*SGVRTSDDKLGALCMQSLQDNVHSIREAAANN*KRLAEE 184
Query: 467 FGPDWAMQ 474
GP WAMQ
Sbjct: 185 CGPAWAMQ 208
>TC228725 homologue to UP|Q9FMF9 (Q9FMF9) Nuclear protein-like, partial (26%)
Length = 1280
Score = 43.1 bits (100), Expect = 3e-04
Identities = 44/191 (23%), Positives = 89/191 (46%), Gaps = 7/191 (3%)
Frame = +2
Query: 364 LSLLKDEFPDVRLNIISKLDQVNQVIGIDLLS---QSLLPAIVELAEDRHWRVRLAIIEY 420
+S+ +E+P+V +I+ L + VIG+ ++ + LLP + + ++RH +V+ I+
Sbjct: 11 MSIXGEEYPEVLGSILGALKSIVNVIGMTKMTPPIKDLLPRLTPILKNRHEKVQENCIDL 190
Query: 421 IPLLASQLGVGFFDDK-LGALC---MQWLKDKVYSIRDAAANNVKRLAEEFGPDWAMQHI 476
+ +A + G F + +C ++ LK IR A N +A+ GP Q +
Sbjct: 191 VGRIADR-GAEFVPAREWMRICFELLEMLKAHKKGIRRATVNTFGYIAKAIGP----QDV 355
Query: 477 IPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAK 536
+ +L+ + R+ AI+++A + +LP ++N + N++ V K
Sbjct: 356 LATLLNNLKVQERQNRVCTTVAIAIVA----ETCSPFTVLPALMNEYRVPELNVQNGVLK 523
Query: 537 VLQSLIPIVEE 547
L L + E
Sbjct: 524 SLSFLFEYIGE 556
>TC227821 weakly similar to UP|Q7PY15 (Q7PY15) EbiP8519 (Fragment), partial
(5%)
Length = 1743
Score = 34.7 bits (78), Expect = 0.12
Identities = 83/358 (23%), Positives = 142/358 (39%), Gaps = 28/358 (7%)
Frame = +3
Query: 102 VRDKSVESLCRIGAQMREQDLVEHFIPLVKRLAA--GEWFTARVSSCGLFHIAY------ 153
+R+++ L + EQ L E IP+ L G+ F +V S L +
Sbjct: 321 LREQAALGLGELIEVTSEQSLKEFVIPITGPLIRIIGDRFPWQVKSAILSTLTTMIKKGG 500
Query: 154 ----PSAPEAQKAELRAMYGQLCQDDMPMVRRSAATNLGKFAA-TVEAAHLKSDIMSVFD 208
P P+ Q ++ + QD VR SAA LGK + + L SD++S
Sbjct: 501 ISLKPFLPQLQTTFVKCL-----QDSTRTVRSSAALALGKLSGLSTRVDPLVSDLLSSLQ 665
Query: 209 DLTQDDQDSVRLLAVEGCAA-LGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYE 267
+D++ L A++G GK L ++ + D RVR ++ L
Sbjct: 666 GSDGGVRDAI-LTALKGVLKHAGKNLS-SAVRTRFYSILKDLIHDDDDRVRTYASSILGI 839
Query: 268 LCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSPELA-------IEHI 320
L + + + TEL+ L + R G + S +L A I
Sbjct: 840 LTQYL-EDVQLTELIQELSSLANSSSWPPR---HGSILTISSLLHYNPATICSSSLFPTI 1007
Query: 321 LPCVKELSADSSQHVR----SALASVIMGMAPVLGKDATI-EQLLPIFLSLLKDEFPDVR 375
+ C+++ D +R AL +++ + V D + + +L + +S D+ +VR
Sbjct: 1008VDCLRDTLKDEKFPLRETSTKALGRLLLYRSQVDPSDTLLYKDVLSLLVSSTHDDSSEVR 1187
Query: 376 LNIISKLDQVNQVIGIDLLSQSLL--PAIVELAEDRHWRVRLAIIEYIPLLASQLGVG 431
+S + V + ++S + PA+ E +D + VRLA E L A QL G
Sbjct: 1188RRALSAIKAVAKANPSAIMSLGTIVGPALAECMKDGNTPVRLA-AERCALHAFQLTKG 1358
>TC216693
Length = 805
Score = 30.4 bits (67), Expect = 2.3
Identities = 31/149 (20%), Positives = 58/149 (38%), Gaps = 1/149 (0%)
Frame = +1
Query: 220 LLAVEGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRT 279
LL V+ + + V+H+LP+IV D R++ V + L + + + +
Sbjct: 178 LLLVKHAEFIINKTSQEHLVSHVLPMIVRAYDDTDARLQEEVLKKSVSLAKQLDAQLVKQ 357
Query: 280 ELVPAYVRL-LRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSA 338
++P L L+ A VR+ A + L ++ + + + D S
Sbjct: 358 VVLPRVHGLALKTTVAAVRVNALLCLGDMVSRLDKHAVLDILQTIQRCTAVDRSPPTLMC 537
Query: 339 LASVIMGMAPVLGKDATIEQLLPIFLSLL 367
V + G + E LLP+ + LL
Sbjct: 538 TLGVANSIFKQYGVEFVAEHLLPLLMPLL 624
>BF069988
Length = 400
Score = 28.9 bits (63), Expect = 6.6
Identities = 33/120 (27%), Positives = 48/120 (39%), Gaps = 10/120 (8%)
Frame = +3
Query: 177 PMVRRSAATNLGKFAATVEAAHL----KSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKL 232
P R AAT LG A + + +M + L + + ++ A AL +
Sbjct: 21 PKDRAEAATELGSLARDNDRTKFIILDEGGVMPLLKLLKEASSPAAQVAAAN---ALVNI 191
Query: 233 LEPQECV------AHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYV 286
Q+ V +H +P+IV D RVR VAN + + E E R E V A V
Sbjct: 192 TTNQDRVVTFIVESHAVPIIVQVLGDSPMRVRVSVANLVSAMAEQ--HELAREEFVRANV 365
>TC231065
Length = 1117
Score = 28.9 bits (63), Expect = 6.6
Identities = 16/50 (32%), Positives = 26/50 (52%)
Frame = +3
Query: 445 LKDKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMT 494
LKDK S +AA N K E +GP M+ + + D+ N+ +Y ++
Sbjct: 726 LKDKSKSHEEAAEVNRKYKEEFYGPTLEMEGCLKSIGDIFNEALAVYNVS 875
>BU761650
Length = 363
Score = 28.9 bits (63), Expect = 6.6
Identities = 12/28 (42%), Positives = 22/28 (77%)
Frame = +1
Query: 287 RLLRDNEAEVRIAAAGKVTKFSRILSPE 314
+LLR ++ +V+IAAA +++ +RI +PE
Sbjct: 253 KLLRHSDDDVKIAAASCISEITRITAPE 336
>TC206007 similar to GB|AAO64828.1|29028898|BT005893 At3g56480 {Arabidopsis
thaliana;} , partial (61%)
Length = 1530
Score = 28.9 bits (63), Expect = 6.6
Identities = 27/117 (23%), Positives = 44/117 (37%), Gaps = 6/117 (5%)
Frame = +2
Query: 154 PSAPEAQKAELRAMYGQLCQDDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQD 213
PS A + ELRA+ Q+ + + ++ + K +D
Sbjct: 449 PSKVMAMEYELRALRDQIREKSIFSIKLQKELTMSK----------------------RD 562
Query: 214 DQDSVRLLAVEGCAALGKLLEPQECVAHILPVI------VNFSQDKSWRVRYMVANQ 264
+++ RL ++G ALG L Q C A + V S + SWR AN+
Sbjct: 563 EENKSRLYMLDGSEALGSYLRVQPCSAEVPQVSKCSFQWYRLSSEGSWREVISGANK 733
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,744,649
Number of Sequences: 63676
Number of extensions: 292413
Number of successful extensions: 1278
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 1242
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1266
length of query: 587
length of database: 12,639,632
effective HSP length: 103
effective length of query: 484
effective length of database: 6,081,004
effective search space: 2943205936
effective search space used: 2943205936
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0310b.14


