

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0310b.11
(371 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
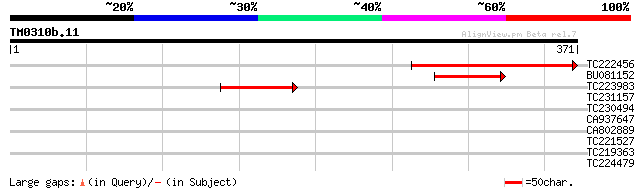
Score E
Sequences producing significant alignments: (bits) Value
TC222456 similar to UP|Q93Z29 (Q93Z29) At1g68600/F24J5_14, parti... 174 4e-44
BU081152 64 8e-11
TC223983 weakly similar to UP|Q93Z29 (Q93Z29) At1g68600/F24J5_14... 59 5e-09
TC231157 similar to GB|CAC82909.1|25136916|ATH297948 cellulose s... 37 0.019
TC230494 homologue to UP|Q8GUZ9 (Q8GUZ9) Cellulose synthase-like... 34 0.093
CA937647 32 0.60
CA802889 30 2.3
TC221527 similar to UP|O81347 (O81347) Plant adhesion molecule 1... 29 3.9
TC219363 similar to UP|Q9FZA1 (Q9FZA1) F3H9.7 protein, partial (... 28 6.6
TC224479 28 8.7
>TC222456 similar to UP|Q93Z29 (Q93Z29) At1g68600/F24J5_14, partial (24%)
Length = 394
Score = 174 bits (442), Expect = 4e-44
Identities = 81/108 (75%), Positives = 90/108 (83%)
Frame = +1
Query: 264 LVAKNFMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQSTSTEDAL 323
LV KNF GVA SLEG VN YL C+ YERVPSKIL YQASDD +Y GYR+A+QS+S E++L
Sbjct: 7 LVVKNFKGVATSLEGCVNGYLQCVAYERVPSKILVYQASDDPLYRGYRAAVQSSSQEESL 186
Query: 324 MGFAVWEPPHGRYKMLKYPWSNYVKVSGALRHCAFMVMAMHGCILSEI 371
+ FA WEPPHG YK YPW +YVKVSGALRHCAFMVMAMHGCILSEI
Sbjct: 187 VDFASWEPPHGPYKTFNYPWRSYVKVSGALRHCAFMVMAMHGCILSEI 330
>BU081152
Length = 426
Score = 64.3 bits (155), Expect = 8e-11
Identities = 27/46 (58%), Positives = 40/46 (86%)
Frame = -3
Query: 279 VVNNYLNCIEYERVPSKILTYQASDDVVYSGYRSALQSTSTEDALM 324
V+N+YL+ +EY+ VPSK LTYQA+DD +Y+G+RS ++STS ED+L+
Sbjct: 163 VINHYLHGVEYKEVPSKFLTYQAADDQIYNGHRSVVESTSKEDSLV 26
>TC223983 weakly similar to UP|Q93Z29 (Q93Z29) At1g68600/F24J5_14, partial
(9%)
Length = 966
Score = 58.5 bits (140), Expect = 5e-09
Identities = 25/50 (50%), Positives = 41/50 (82%)
Frame = +1
Query: 139 FSIGATLSKGLNRGLGTLTAGGLAVGMGVLSKLAGQWEEIIVMVSIFIVG 188
+ +GATL+KG NR LGT++AGGLA+G+ L+ L+G++EE+I++++ I G
Sbjct: 787 YFVGATLNKGFNRSLGTISAGGLALGIAELAVLSGKFEELIIVLNXXIAG 936
>TC231157 similar to GB|CAC82909.1|25136916|ATH297948 cellulose synthase-like
protein {Arabidopsis thaliana;} , partial (23%)
Length = 932
Score = 36.6 bits (83), Expect = 0.019
Identities = 20/56 (35%), Positives = 32/56 (56%), Gaps = 1/56 (1%)
Frame = +1
Query: 209 VFLITYCYIIVSGYQTGKFIDTAVS-RFLLIALGAAVSLGVNVCLYPIWAGEDLHD 263
+FLI YC++ +G+FI ++S FL+ LG ++L + L W+G LHD
Sbjct: 166 MFLIVYCFLPAVSLFSGQFIVQSLSATFLVFLLGITITLCLLALLEIKWSGITLHD 333
>TC230494 homologue to UP|Q8GUZ9 (Q8GUZ9) Cellulose synthase-like protein D4
(Fragment), partial (20%)
Length = 769
Score = 34.3 bits (77), Expect = 0.093
Identities = 27/102 (26%), Positives = 43/102 (41%), Gaps = 5/102 (4%)
Frame = +1
Query: 209 VFLITYCYIIVSGYQTGKFI-DTAVSRFLLIALGAAVSLGVNVCLYPIWAGEDLHDLVAK 267
+FLI YC++ +G+FI T FL LG V+L + L W+G +L +
Sbjct: 13 IFLIVYCFLPALSXFSGQFIVQTLNVTFLSYLLGITVTLCMLAVLEIKWSGIELEEWWRN 192
Query: 268 N----FMGVAASLEGVVNNYLNCIEYERVPSKILTYQASDDV 305
G +A L V+ L + + + + DDV
Sbjct: 193 EQFWLIGGTSAHLAAVLQGLLKVVAGIEISFTLTSKSGGDDV 318
>CA937647
Length = 391
Score = 31.6 bits (70), Expect = 0.60
Identities = 19/53 (35%), Positives = 24/53 (44%)
Frame = -1
Query: 34 GYSQIGIPLPESDDDNDGPPGSKNCCRFCSYRALSDRLVGAWKTTKRVAARAW 86
G + +G L DDD+DG G C C +SD V W T +V AW
Sbjct: 181 GGALMGGMLKFLDDDDDGGCGGCGSCDACF--VVSDEPVSIWTTCLKVEVSAW 29
>CA802889
Length = 457
Score = 29.6 bits (65), Expect = 2.3
Identities = 26/78 (33%), Positives = 40/78 (50%), Gaps = 4/78 (5%)
Frame = +1
Query: 180 VMVSIFIVGFCATYAKL-YPTMKAYEYGFRVFLITYCYIIVSGYQTGKFIDTAVSRFLLI 238
V+ ++F+V C + A L Y + E+GF + ++IVS + +D LL+
Sbjct: 205 VVGNVFVV--C*SMAILVYFILLMVEFGFG*EMCYCWHLIVSFFNV*VLVDVHAEPNLLV 378
Query: 239 ALGAA---VSLGVNVCLY 253
A G A SLG+ VCLY
Sbjct: 379 AYGKA*LLNSLGLYVCLY 432
>TC221527 similar to UP|O81347 (O81347) Plant adhesion molecule 1 (PAM1),
partial (41%)
Length = 638
Score = 28.9 bits (63), Expect = 3.9
Identities = 18/42 (42%), Positives = 25/42 (58%)
Frame = +1
Query: 81 VAARAWEMGLSDPRKIVFAAKMGLALILISLLIFLKTPFEDL 122
+A R W++ L + KIVF K+GLAL+ +K PFE L
Sbjct: 220 LALRIWDVFLYEGVKIVF--KVGLALLKYCHDDLIKLPFEKL 339
>TC219363 similar to UP|Q9FZA1 (Q9FZA1) F3H9.7 protein, partial (44%)
Length = 1156
Score = 28.1 bits (61), Expect = 6.6
Identities = 23/73 (31%), Positives = 29/73 (39%), Gaps = 11/73 (15%)
Frame = -3
Query: 67 LSDRLVGAWKTTKRVAARAWEM----GLSDPRKIVFAAKMG-------LALILISLLIFL 115
+SD V W T +V AW GL +V + G L L+LI LL L
Sbjct: 431 VSDEPVSIWTTCLKVEVSAWTKVVG*GLESDLGMVLGVRGGVGIWMPLLLLLLIQLLTLL 252
Query: 116 KTPFEDLGRHSVW 128
F GR +W
Sbjct: 251 VGEFIGEGRLWLW 213
>TC224479
Length = 497
Score = 27.7 bits (60), Expect = 8.7
Identities = 20/58 (34%), Positives = 25/58 (42%)
Frame = -2
Query: 103 GLALILISLLIFLKTPFEDLGRHSVWAILTVVTVFEFSIGATLSKGLNRGLGTLTAGG 160
GL +L+ L K+PF L A+L +F I TLS G LG GG
Sbjct: 277 GLGALLLELCF--KSPFHCLTGRGS*ALLGYPEIFRLLIFGTLSSGAAEKLGPPVLGG 110
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.139 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,667,150
Number of Sequences: 63676
Number of extensions: 248633
Number of successful extensions: 1584
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1570
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1579
length of query: 371
length of database: 12,639,632
effective HSP length: 99
effective length of query: 272
effective length of database: 6,335,708
effective search space: 1723312576
effective search space used: 1723312576
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0310b.11


