

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
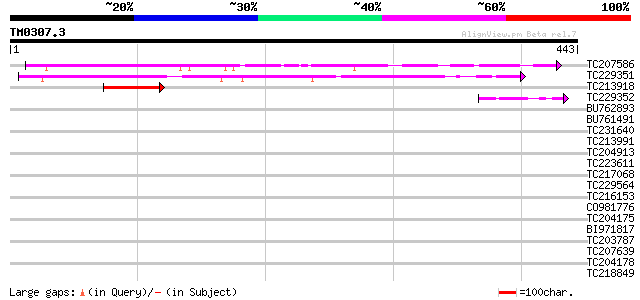
Query= TM0307.3
(443 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207586 similar to GB|BAD10504.1|42409240|AP005544 auxin-regula... 173 1e-43
TC229351 similar to GB|AAP37875.1|30725706|BT008516 At5g59790 {A... 165 3e-41
TC213918 similar to UP|Q9SYJ8 (Q9SYJ8) F3F20.2 protein, partial ... 60 2e-09
TC229352 similar to GB|AAP37875.1|30725706|BT008516 At5g59790 {A... 46 3e-05
BU762893 similar to GP|15808946|gb| auxin-regulated protein {Lyc... 40 0.002
BU761491 40 0.003
TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated anti... 36 0.030
TC213991 similar to UP|Q6H976 (Q6H976) STYLOSA protein, partial ... 36 0.030
TC204913 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, ... 33 0.34
TC223611 weakly similar to UP|Q9LJ56 (Q9LJ56) Gb|AAD43149.1, par... 32 0.57
TC217068 similar to UP|Q93Z30 (Q93Z30) AT4g18950/F13C5_120 (Prot... 31 1.3
TC229564 weakly similar to UP|Q6VSS7 (Q6VSS7) TOX, partial (18%) 31 1.3
TC216153 similar to UP|Q9LLM3 (Q9LLM3) MTD1, partial (61%) 31 1.3
CO981776 31 1.3
TC204175 homologue to UP|TBB2_SOYBN (P12460) Tubulin beta-2 chai... 30 1.7
BI971817 30 1.7
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 30 1.7
TC207639 similar to UP|O48592 (O48592) GT2 protein, partial (24%) 30 2.2
TC204178 homologue to UP|TBB7_MAIZE (Q41784) Tubulin beta-7 chai... 30 2.8
TC218849 similar to UP|Q705X4 (Q705X4) Rho GDP dissociation inhi... 30 2.8
>TC207586 similar to GB|BAD10504.1|42409240|AP005544 auxin-regulated
protein-like {Oryza sativa (japonica cultivar-group);} ,
partial (40%)
Length = 1981
Score = 173 bits (439), Expect = 1e-43
Identities = 155/470 (32%), Positives = 219/470 (45%), Gaps = 51/470 (10%)
Frame = +3
Query: 13 SRDTSPDRAKICRKV---IKPARKVQVVYYLFRNGLLEHPHFMELTLLPNQPLRLKDVFD 69
+R SP RAK+ + + KV V+YYL RN LEHPHFME+ L L L+DV D
Sbjct: 183 NRQVSPVRAKVWTEKSPKYHQSLKVPVIYYLSRNRQLEHPHFMEVPLSSPDGLYLRDVID 362
Query: 70 RLMVLRGSGMPLQYSWSSKRNYKSGYVWFDLALKDVIYPSEGGEYVLKGSELV-EGCSER 128
RL VLRG M YSWS KR+YKSG+VW DL D+I P+ G EYVLKGSEL E S+R
Sbjct: 363 RLNVLRGRSMASLYSWSCKRSYKSGFVWHDLCEDDIILPAHGNEYVLKGSELFDESNSDR 542
Query: 129 FQQL------------------------NVSNKNG----IHEPPETNYGYNAKSKVLSNR 160
F + + S+ NG I + E + + S +S
Sbjct: 543 FSPISNVKTQSVKLLPGPASSRSLDEGSSSSSMNGKETRISQDDELSQDPHTGSSDVSPE 722
Query: 161 QQQKEAE-------EYEEYE--------EQEEEKEFTSSTTTPHSRCSRGVSTEELGEDE 205
+ ++++ EY+ Y+ Q EEK+ + + C+RGVSTE+ +
Sbjct: 723 SRAEKSDALSLALTEYKIYKTDGLADASTQTEEKD---NRSRAQKTCTRGVSTEDGSLES 893
Query: 206 DRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQ 265
+ HE Q + +V T + R V+ PPP + S++ AE L +
Sbjct: 894 ECHEICQ--AEAPQVKDTPRI-CRDAVS--PPPSTSSSSSFVGKAETLESLIRADASKVN 1058
Query: 266 RFG--GELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTAS-KQRQ 322
F E + T+ + L+QLI+CGS + S G S K R
Sbjct: 1059SFRILEEESMQMPTNTRMKASNLLMQLISCGSI----------SVKNHSFGLIPSYKPRF 1208
Query: 323 SVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESM 382
S + F S V + ++ENP+LM L E+KEYFSGSLVE +
Sbjct: 1209SSSKFPSPLFSTS--------FVLGEFDCLAENPKLMSLRL---EDKEYFSGSLVETKLK 1355
Query: 383 KAN-HPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEKCIP 431
+ + H VLKRS+SYN+ER K++ ++E++E KCIP
Sbjct: 1356EGDGHNVLKRSSSYNDER---------TFKEQKQQEDKEESSSGHSKCIP 1478
>TC229351 similar to GB|AAP37875.1|30725706|BT008516 At5g59790 {Arabidopsis
thaliana;} , partial (58%)
Length = 1212
Score = 165 bits (418), Expect = 3e-41
Identities = 135/418 (32%), Positives = 197/418 (46%), Gaps = 22/418 (5%)
Frame = +3
Query: 8 PHRCSSRDTSPDRAKIC---RKVIKPARKVQVVYYLFRNGLLEHPHFMELTLLPNQPLRL 64
P + + R+TSP+R K+ + K RKV VVYYL RNG LEHPHFME+ L L L
Sbjct: 63 PKKWTERETSPERTKVWAEPKPKPKTPRKVSVVYYLSRNGQLEHPHFMEVPLSSPHGLYL 242
Query: 65 KDVFDRLMVLRGSGMPLQYSWSSKRNYKSGYVWFDLALKDVIYPSEGG-EYVLKGSELVE 123
KDV +RL LRG GM YSWS+KR+YK+G+VW DL+ D IYP+ G +Y+LKGSE+VE
Sbjct: 243 KDVINRLNALRGKGMATLYSWSAKRSYKNGFVWHDLSENDFIYPTTXGQDYILKGSEIVE 422
Query: 124 GCSERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQK----EAEEYEEYEEQEEEK 179
N G + + V++ R+ Q + EY Y+ +
Sbjct: 423 -----------HNVTGAGARVNKSEEESDSPVVITRRRNQSWSSIDMNEYRVYKSESFGD 569
Query: 180 E-----FTSSTTTPHSRCSRGVSTE-ELGEDEDRHEALQNNTKTEKVV-VTNNVSSRGNV 232
++T T R R E E E +++++ ++ + E+V VT + + +
Sbjct: 570 SAGRIGADAATQTEEKRKRRRAGREGEAVEIQEKNDGIEAGMEGERVAHVTCDDDNNNHT 749
Query: 233 TNV------PPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSA 286
T + PPP + +L R + ++S + L ES S S
Sbjct: 750 TELSREEISPPPSDSSPETLETLMRADGR---LGLRSSESEKENLTVESCPSGRTRASSV 920
Query: 287 LLQLIACGS-SGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAV 345
LLQL++CG+ S E A ++ S VG S+ + S E +
Sbjct: 921 LLQLLSCGAVSFKECGANAVKDQGFSLVGHYKSRMPRGAGNHSGNETGTSMEIPD----- 1085
Query: 346 AMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKL 403
++RV E+KEYFSGSL+ E+ K P LKRS+SYN + S+L
Sbjct: 1086---LSRV------------RLEDKEYFSGSLI--ETKKVETPALKRSSSYNADSGSRL 1208
>TC213918 similar to UP|Q9SYJ8 (Q9SYJ8) F3F20.2 protein, partial (14%)
Length = 569
Score = 60.5 bits (145), Expect = 2e-09
Identities = 28/48 (58%), Positives = 33/48 (68%)
Frame = +2
Query: 74 LRGSGMPLQYSWSSKRNYKSGYVWFDLALKDVIYPSEGGEYVLKGSEL 121
LRG +P +SWS KR YKSGYVW DL D+I P EYVLKGS++
Sbjct: 17 LRGKDLPDAFSWSYKRRYKSGYVWQDLLDDDLITPISDNEYVLKGSQI 160
>TC229352 similar to GB|AAP37875.1|30725706|BT008516 At5g59790 {Arabidopsis
thaliana;} , partial (13%)
Length = 825
Score = 46.2 bits (108), Expect = 3e-05
Identities = 29/70 (41%), Positives = 39/70 (55%)
Frame = +1
Query: 367 EEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVK 426
E+KEYFSGSL+E + K P LKRS+SYN + S+L + +E E V+
Sbjct: 166 EDKEYFSGSLIETK--KVETPALKRSSSYNADSGSRLQI--------VEHEGE----AVR 303
Query: 427 EKCIPIKKSS 436
KCIP K +
Sbjct: 304 AKCIPRKSKT 333
>BU762893 similar to GP|15808946|gb| auxin-regulated protein {Lycopersicon
esculentum}, partial (6%)
Length = 418
Score = 40.0 bits (92), Expect = 0.002
Identities = 20/34 (58%), Positives = 24/34 (69%), Gaps = 1/34 (2%)
Frame = +2
Query: 33 KVQVVYYLFRNGLLEHPHFMELTL-LPNQPLRLK 65
KV V+YYL RNG LEHPH ME+ + P + L LK
Sbjct: 317 KVPVIYYLSRNGQLEHPHLMEVPISSPQRVLCLK 418
>BU761491
Length = 433
Score = 39.7 bits (91), Expect = 0.003
Identities = 21/50 (42%), Positives = 29/50 (58%), Gaps = 3/50 (6%)
Frame = +2
Query: 5 QVRPHRCSSRDTSPDRAKICRKV---IKPARKVQVVYYLFRNGLLEHPHF 51
+ R + +R SP+RAK+ + + KV V+YYL RN LEHPHF
Sbjct: 284 EARMKKKYNRQVSPERAKVWTEKSPKYHQSLKVPVIYYLCRNRQLEHPHF 433
>TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated antigen, partial
(7%)
Length = 466
Score = 36.2 bits (82), Expect = 0.030
Identities = 24/71 (33%), Positives = 36/71 (49%), Gaps = 5/71 (7%)
Frame = +1
Query: 156 VLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGE-----DEDRHEA 210
VLS+ Q+++E +E EE EE+EEE+E + EE G DED+H+
Sbjct: 7 VLSDTQEEEEEKEEEEEEEEEEEEE----EEEEEEEEEKEEEEEEEGNVLVIIDEDKHDT 174
Query: 211 LQNNTKTEKVV 221
+ N K E +
Sbjct: 175 EELNKKCEDFI 207
>TC213991 similar to UP|Q6H976 (Q6H976) STYLOSA protein, partial (12%)
Length = 440
Score = 36.2 bits (82), Expect = 0.030
Identities = 41/166 (24%), Positives = 70/166 (41%), Gaps = 17/166 (10%)
Frame = +3
Query: 160 RQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEK 219
+QQQ++ ++ ++ ++Q+++ + + P + SR DR L
Sbjct: 18 QQQQQQQQQQQQQQQQQQQPQSQQQQSQPQQQQSR-----------DRAHLL-------- 140
Query: 220 VVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKL-NRRVVVPVQS--------KQRFGGE 270
N S+ G V N A +LA K+ R+ +P+Q KQRFG
Sbjct: 141 -----NGSANGLVGN--------PGTANALATKMYEERLKLPLQRDPLDDAAMKQRFGEN 281
Query: 271 LG--------TESATSAAPSRYSALLQLIACGSSGPEFKARTQQEP 308
+G + ++AAP + S + A G P+ +ARTQQ P
Sbjct: 282 MGQLLDPNHASILKSAAAPGQPSGQVLHGAAGGMSPQVQARTQQLP 419
>TC204913 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, partial
(14%)
Length = 1080
Score = 32.7 bits (73), Expect = 0.34
Identities = 44/164 (26%), Positives = 62/164 (36%), Gaps = 20/164 (12%)
Frame = +3
Query: 212 QNNTKTEKVVVTNNVSSRGNVTNVPPPMK-----NHSAAAASLAEKLNRRVVVPVQSKQR 266
+N K+ K VVT N + +T+V PP + A ++ E L V++P+Q +
Sbjct: 207 KNMKKSLKDVVTENEFEKKLLTDVIPPTDIGVTFDDIGALENVKETLKELVMLPLQRPEL 386
Query: 267 FG--------------GELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSS 312
FG G GT A A I S K + E V +
Sbjct: 387 FGKGQLAKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKA 566
Query: 313 VGTTASKQRQSVDRRDQKAFMMSCEEEE-EEEAVAMMMNRVSEN 355
V + ASK SV D+ M+ E E EA+ M N N
Sbjct: 567 VFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVN 698
>TC223611 weakly similar to UP|Q9LJ56 (Q9LJ56) Gb|AAD43149.1, partial (12%)
Length = 463
Score = 32.0 bits (71), Expect = 0.57
Identities = 27/114 (23%), Positives = 45/114 (38%), Gaps = 14/114 (12%)
Frame = +2
Query: 119 SELVEGCSE--RFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQ------------QK 164
SEL+ E R LN + + E ++G + + R+Q Q+
Sbjct: 89 SELIGRLREFLRSSDLNTTTTATVRRQLEADFGIDLSDRKAFIREQVDLFLQTEHNQPQQ 268
Query: 165 EAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTE 218
E + ++ EEQEE+ + P EE E+ED+ E +N K +
Sbjct: 269 EERQNDDVEEQEEDAPNNPEQSQPSDSKEETDEEEEGEEEEDKPEQAKNAKKNK 430
>TC217068 similar to UP|Q93Z30 (Q93Z30) AT4g18950/F13C5_120 (Protein
kinase-like protein), partial (88%)
Length = 1650
Score = 30.8 bits (68), Expect = 1.3
Identities = 32/97 (32%), Positives = 47/97 (47%), Gaps = 11/97 (11%)
Frame = +2
Query: 154 SKVLSNRQQQKEAEEYEEYEEQEEEKEFTSST-TTPHSRCS---RG--VSTEELGE---- 203
+K L A E EYE +E +FT+S T + CS RG V+ ++LGE
Sbjct: 371 AKPLMAPMHVNHAREVPEYEINPKELDFTNSVEITKGTFCSALRRGTKVAVKKLGEDVIS 550
Query: 204 DEDRHEALQNNTKT-EKVVVTNNVSSRGNVTNVPPPM 239
DE++ +AL++ +K+ N V G VT P M
Sbjct: 551 DEEKVKALRDELALFQKIRHPNVVQFLGAVTQSSPMM 661
>TC229564 weakly similar to UP|Q6VSS7 (Q6VSS7) TOX, partial (18%)
Length = 609
Score = 30.8 bits (68), Expect = 1.3
Identities = 20/75 (26%), Positives = 34/75 (44%), Gaps = 4/75 (5%)
Frame = +3
Query: 160 RQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEK 219
+++ KEAE+ E E + EEK+ + G E GE+++ + + +K
Sbjct: 135 KEESKEAEKPAEEENKPEEKKEEEKPPEEPKKEEGGEGGEGGGENKEEKGGEEGKEEAKK 314
Query: 220 ----VVVTNNVSSRG 230
VVV NN+ G
Sbjct: 315 EDNVVVVANNIDDEG 359
>TC216153 similar to UP|Q9LLM3 (Q9LLM3) MTD1, partial (61%)
Length = 1021
Score = 30.8 bits (68), Expect = 1.3
Identities = 33/138 (23%), Positives = 53/138 (37%), Gaps = 7/138 (5%)
Frame = +2
Query: 306 QEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAVAM-----MMNRVSENPRLML 360
QE T+ S S+ R + S EE E E A M + E +
Sbjct: 134 QEEEEEECSTSTSS---SIGRNSDVSSERSMEEGENEVESAYHGPLHAMETLEEVLPIRR 304
Query: 361 G--NLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEEN 418
G N N + K + + L + S + + K N+Y RR+ + + V K
Sbjct: 305 GISNFYNGKSKSFTT--LADAVSSPSVKDIAKPENAYTRRRRNLMALNHVLDKNNRNYPL 478
Query: 419 RERKGGVKEKCIPIKKSS 436
R GG+ ++ I + +SS
Sbjct: 479 RSSGGGICKRSISLSRSS 532
>CO981776
Length = 714
Score = 30.8 bits (68), Expect = 1.3
Identities = 32/117 (27%), Positives = 48/117 (40%), Gaps = 6/117 (5%)
Frame = -1
Query: 299 EFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCE-EEEEEEAVAMMMNRVSENPR 357
+ + RT++E + SVG S R+S + CE E E E + + S +
Sbjct: 669 QLRGRTRRETKXRSVGEIQSGLRRSKRATKNRINYRQCEASESETEFIKSEKSNSSADHS 490
Query: 358 LMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNE-----ERRSKLGVEEVK 409
N + E E S E + MK + PV + NE E+ S G EEV+
Sbjct: 489 DPNENGEYMMESEDSDDSDNEEQEMKVDDPVTYPAVEENEQNQPPEKLSSPGQEEVE 319
>TC204175 homologue to UP|TBB2_SOYBN (P12460) Tubulin beta-2 chain, complete
Length = 1653
Score = 30.4 bits (67), Expect = 1.7
Identities = 16/43 (37%), Positives = 22/43 (50%)
Frame = +1
Query: 139 GIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEF 181
G+ E T N V +Q Q + +EYEE+EEE+EF
Sbjct: 1276 GMDEMEFTEAESNMNDLVSEYQQYQDATADEDEYEEEEEEEEF 1404
>BI971817
Length = 472
Score = 30.4 bits (67), Expect = 1.7
Identities = 31/122 (25%), Positives = 47/122 (38%)
Frame = +2
Query: 177 EEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVP 236
EEK T+ T P + CS EE + + AL +T ++ T NV
Sbjct: 74 EEKSSTAVLTMPETHCSPQCKDEESSDCDKACNALLPSTSIDQHQTT-------GCRNVD 232
Query: 237 PPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSS 296
+ H + SL EKL+ R+ S+ L + S S ++L+ C S
Sbjct: 233 VNDQTHLGSQVSL-EKLDSRIDSKSTSRVPTSSTLLCQEMNSTGSSLKVSVLEQEQCRES 409
Query: 297 GP 298
P
Sbjct: 410 KP 415
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 30.4 bits (67), Expect = 1.7
Identities = 22/96 (22%), Positives = 42/96 (42%)
Frame = -3
Query: 133 NVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRC 192
N +NK+ + PE G + N ++ ++ + ++ ++ E++ +
Sbjct: 520 NSNNKSNSKKAPEGGAGGAEE-----NGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEED 356
Query: 193 SRGVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSS 228
G EE EDE+ EALQ K +K + + SS
Sbjct: 355 EDGAEDEENEEDEEDEEALQPPKKRKK*I*FSKASS 248
>TC207639 similar to UP|O48592 (O48592) GT2 protein, partial (24%)
Length = 1689
Score = 30.0 bits (66), Expect = 2.2
Identities = 16/38 (42%), Positives = 22/38 (57%)
Frame = +1
Query: 143 PPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKE 180
PP AK + R++++E EE EE EE+EEE E
Sbjct: 862 PPSQYEQGAAKENNNNERKEREEEEEDEEEEEEEEEDE 975
>TC204178 homologue to UP|TBB7_MAIZE (Q41784) Tubulin beta-7 chain (Beta-7
tubulin), complete
Length = 1720
Score = 29.6 bits (65), Expect = 2.8
Identities = 16/42 (38%), Positives = 21/42 (49%)
Frame = +3
Query: 139 GIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKE 180
G+ E T N V +Q Q + EEYEE+EEE+E
Sbjct: 1350 GMDEMEFTEAESNMNDLVAEYQQYQDATADEEEYEEEEEEEE 1475
>TC218849 similar to UP|Q705X4 (Q705X4) Rho GDP dissociation inhibitor 1,
partial (79%)
Length = 826
Score = 29.6 bits (65), Expect = 2.8
Identities = 24/67 (35%), Positives = 33/67 (48%), Gaps = 10/67 (14%)
Frame = +2
Query: 149 GYNAKSKVLSNRQQ--------QKEAEEYEEYEEQEEEK--EFTSSTTTPHSRCSRGVST 198
G ++ S + S +QQ +KEAEE EE EE+EEE + + T P S
Sbjct: 134 GPSSSSGIASTKQQNPPETFHHRKEAEEEEETEEEEEEDVCQHKNITFVPGPLLSLKDQI 313
Query: 199 EELGEDE 205
E+ EDE
Sbjct: 314 EKDKEDE 334
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.310 0.126 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,704,968
Number of Sequences: 63676
Number of extensions: 216812
Number of successful extensions: 1309
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1259
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1293
length of query: 443
length of database: 12,639,632
effective HSP length: 100
effective length of query: 343
effective length of database: 6,272,032
effective search space: 2151306976
effective search space used: 2151306976
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0307.3


