

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
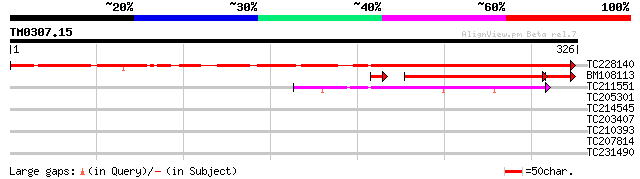
Query= TM0307.15
(326 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC228140 261 3e-70
BM108113 100 2e-24
TC211551 similar to UP|Q9FLD0 (Q9FLD0) Gb|AAD32815.2, partial (13%) 50 1e-06
TC205301 similar to UP|Q7QIP8 (Q7QIP8) AgCP3364 (Fragment), part... 32 0.51
TC214545 GB|AAA33978.1|169989|SOYIDH NADPH-specific isocitrate d... 31 0.66
TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-pr... 30 1.5
TC210393 weakly similar to GB|AAF25189.1|6694227|AF187147 drebri... 28 4.3
TC207814 similar to UP|Q75XU7 (Q75XU7) RIO kinase, partial (43%) 27 9.6
TC231490 similar to UP|HSF8_LYCPE (P41153) Heat shock factor pro... 27 9.6
>TC228140
Length = 1467
Score = 261 bits (667), Expect = 3e-70
Identities = 164/334 (49%), Positives = 210/334 (62%), Gaps = 9/334 (2%)
Frame = +1
Query: 1 MSSEDNFSDGVVKPQPRPVLSDVTNLRAKRPFSSISADDGDSRFTKKKVQSKLGVQ-AQQ 59
M S DNF DG+ K RP L+DVTNL AKR FS +S D GD +FTKK +LGV+ +
Sbjct: 340 MGSNDNFGDGIFKT--RPHLADVTNLPAKRSFSLVSGDGGDLQFTKK---IRLGVENLAK 504
Query: 60 GRSNK--------DKVVVSQQKGQNPCLELPFCGDDSLGNVSSQESNLQLPSGGLEEKNL 111
G+S + V+ Q K ++ L LPF DD+L + N L G+EE+N
Sbjct: 505 GKSQMQFGAHAYLNNEVLLQPKEKHTIL-LPF-SDDTLHSFQ----NPSLTVEGMEEQN- 663
Query: 112 LGGAVVSGPTEVPVAVLGEIGQGRPLNEGFIDRGIESGQRDVRAVDNLGSPKCGGRAVEL 171
P ++ G+ G G DR +ESG +D+ V+NLGSPKCG A ++
Sbjct: 664 --------PLDLENFKFGKEGDGIVAT----DRAVESGGKDICDVENLGSPKCG--AEQM 801
Query: 172 PTMSDSCDSRFPGLERCSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHYQDVK 231
PT+S S DS F GL+ CS +S AADL KSCTCSFCSKA +IWSDLHYQD K
Sbjct: 802 PTISVSNDSNFLGLKPCS--------SSVTEAADL-KSCTCSFCSKAGYIWSDLHYQDAK 954
Query: 232 GRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFA 291
GRLSA+KKSQK+A +QK S ++++VM DQ S ES KLEL+L+HQWKSLF M+N +
Sbjct: 955 GRLSAIKKSQKEAKMIIQKFSGLENTVMHDQHRSEESLKLELSLVHQWKSLFLQMQNMYT 1134
Query: 292 EESRQLESSFETLKDLRDNCKNDLDSTDNTHFDN 325
+ES QLE+SFETLK+LR+NCK DL+ DN+H N
Sbjct: 1135QESSQLETSFETLKNLRENCKTDLELNDNSHHQN 1236
>BM108113
Length = 568
Score = 100 bits (248), Expect(3) = 2e-24
Identities = 49/82 (59%), Positives = 64/82 (77%)
Frame = +3
Query: 228 QDVKGRLSALKKSQKDASNAVQKLSEIKDSVMPDQQSSSESSKLELALMHQWKSLFDFME 287
+D KGRLSA+KKSQK+A +QK S ++D++M DQ S ES KLE +L+HQWKSLF ++
Sbjct: 57 KDAKGRLSAIKKSQKEAKMIIQKFSGLEDTIMHDQHRSEESLKLEFSLVHQWKSLFLEIQ 236
Query: 288 NTFAEESRQLESSFETLKDLRD 309
N + +ES QLESSFETLK LR+
Sbjct: 237 NMYTQESSQLESSFETLKILRE 302
Score = 27.7 bits (60), Expect(3) = 2e-24
Identities = 10/10 (100%), Positives = 10/10 (100%)
Frame = +2
Query: 208 KSCTCSFCSK 217
KSCTCSFCSK
Sbjct: 29 KSCTCSFCSK 58
Score = 22.3 bits (46), Expect(3) = 2e-24
Identities = 7/17 (41%), Positives = 11/17 (64%)
Frame = +1
Query: 309 DNCKNDLDSTDNTHFDN 325
+ CK L+ DN+H +N
Sbjct: 298 EKCKTGLELNDNSHCEN 348
>TC211551 similar to UP|Q9FLD0 (Q9FLD0) Gb|AAD32815.2, partial (13%)
Length = 895
Score = 50.1 bits (118), Expect = 1e-06
Identities = 45/161 (27%), Positives = 73/161 (44%), Gaps = 13/161 (8%)
Frame = +3
Query: 164 CGGRAVELPTMSDSC--DSRFPGLERCSVLQGNAGPTSAAAAADLLKSCTCSFCSKAAHI 221
C R L S S +S+ G +R S G+ G DL + S + A
Sbjct: 123 CNSRTASLLLSSSSSPSNSKIVGDKRGSSHAGH-GARKRVKMKDL-DAVVHSVAATVASQ 296
Query: 222 WSDLHYQDVKGRLSALKKSQKDASNAV---------QKLSEIKDSVMPDQQSSSESSKLE 272
W +L QD+KGRLSAL++S++ + + ++ +D + S+ +
Sbjct: 297 WLELLQQDIKGRLSALRRSRRKVRSVITTELPFLLSKEFGNNQDYDPCTVEMSAGLPTSK 476
Query: 273 LALMH--QWKSLFDFMENTFAEESRQLESSFETLKDLRDNC 311
+A MH +W SLFD M+ +EE +QLE +K+ + C
Sbjct: 477 IADMHRARWSSLFDHMDAALSEEEKQLECWLNQVKEKQLLC 599
>TC205301 similar to UP|Q7QIP8 (Q7QIP8) AgCP3364 (Fragment), partial (10%)
Length = 1199
Score = 31.6 bits (70), Expect = 0.51
Identities = 28/111 (25%), Positives = 41/111 (36%)
Frame = -3
Query: 194 NAGPTSAAAAADLLKSCTCSFCSKAAHIWSDLHYQDVKGRLSALKKSQKDASNAVQKLSE 253
+AGP+ A C FC IWSD Q++ R ++L D + S
Sbjct: 711 SAGPSQKLEQAPQQVQC---FCFSVQQIWSDQTPQELPQRETSLMHRSGDHHKSA---SL 550
Query: 254 IKDSVMPDQQSSSESSKLELALMHQWKSLFDFMENTFAEESRQLESSFETL 304
++S PD + E + H +L S ESS+ TL
Sbjct: 549 FQNSSFPDHTNHIELFGYHMLQFHVHNNLLILKVQLHLPTSLSQESSYNTL 397
>TC214545 GB|AAA33978.1|169989|SOYIDH NADPH-specific isocitrate dehydrogenase
{Glycine max;} , complete
Length = 1727
Score = 31.2 bits (69), Expect = 0.66
Identities = 26/72 (36%), Positives = 37/72 (51%), Gaps = 11/72 (15%)
Frame = -2
Query: 19 VLSDVTNLRAKRPFSSISAD-----DGDSRFTK---KKVQSKLGVQAQQGRSNKDKVVVS 70
VL VT+ R R F+ +SA+ + S+ K +K+ KLGV Q KDK V +
Sbjct: 1690 VLIMVTH*RLGRMFNFLSAERHLQNNNKSKHAK*IQQKIHYKLGVGRDQ*EITKDKSVHN 1511
Query: 71 QQKGQ---NPCL 79
Q G+ NPC+
Sbjct: 1510 QYYGEETANPCI 1475
>TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-protein
(AtAGP4) (AT5g10430/F12B17_220), partial (50%)
Length = 966
Score = 30.0 bits (66), Expect = 1.5
Identities = 14/28 (50%), Positives = 17/28 (60%)
Frame = -1
Query: 113 GGAVVSGPTEVPVAVLGEIGQGRPLNEG 140
G A+ GP E P VLGE+G G + EG
Sbjct: 513 GSALPEGPGEGPGGVLGEVGLGVTVGEG 430
>TC210393 weakly similar to GB|AAF25189.1|6694227|AF187147 drebrin A {Mus
musculus;} , partial (4%)
Length = 784
Score = 28.5 bits (62), Expect = 4.3
Identities = 17/49 (34%), Positives = 28/49 (56%), Gaps = 1/49 (2%)
Frame = +3
Query: 12 VKPQPRPVLSDVTNLRAKRPFSSISADDG-DSRFTKKKVQSKLGVQAQQ 59
++P PRPV +++ + KR +S G R TKKK+ S+ +Q Q+
Sbjct: 483 LRPTPRPVTANLAPVSHKRASTSRDQVQGIKERDTKKKIWSR**IQLQK 629
>TC207814 similar to UP|Q75XU7 (Q75XU7) RIO kinase, partial (43%)
Length = 1259
Score = 27.3 bits (59), Expect = 9.6
Identities = 14/34 (41%), Positives = 17/34 (49%)
Frame = +2
Query: 224 DLHYQDVKGRLSALKKSQKDASNAVQKLSEIKDS 257
DL+YQ + G AL +Q QK S KDS
Sbjct: 539 DLYYQTITGLKHALSLTQPSQQKTQQKSSPTKDS 640
>TC231490 similar to UP|HSF8_LYCPE (P41153) Heat shock factor protein HSF8
(Heat shock transcription factor 8) (HSTF 8) (Heat
stress transcription factor), partial (32%)
Length = 646
Score = 27.3 bits (59), Expect = 9.6
Identities = 16/43 (37%), Positives = 19/43 (43%), Gaps = 1/43 (2%)
Frame = -1
Query: 174 MSDSCDSRFPGLERCSVLQGNAG-PTSAAAAADLLKSCTCSFC 215
++ SC S F L R S + PTS A D L C C C
Sbjct: 607 LTSSCMSTFLSLLRISTSSSSPNFPTSTQAPTDELCPCACCAC 479
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.129 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,331,398
Number of Sequences: 63676
Number of extensions: 173639
Number of successful extensions: 713
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 709
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 709
length of query: 326
length of database: 12,639,632
effective HSP length: 98
effective length of query: 228
effective length of database: 6,399,384
effective search space: 1459059552
effective search space used: 1459059552
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0307.15


