

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0306.4
(968 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
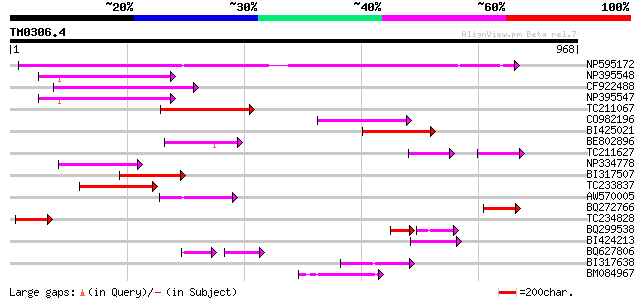
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 630 0.0
NP395548 reverse transcriptase [Glycine max] 154 2e-37
CF922488 150 2e-36
NP395547 reverse transcriptase [Glycine max] 144 2e-34
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 144 2e-34
CO982196 115 7e-26
BI425021 110 3e-24
BE802896 108 1e-23
TC211627 70 2e-22
NP334778 reverse transcriptase [Glycine max] 104 2e-22
BI317507 101 1e-21
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 98 2e-20
AW570005 93 5e-19
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 93 5e-19
TC234828 91 2e-18
BQ299538 60 2e-17
BI424213 84 3e-16
BQ627806 56 9e-14
BI317638 weakly similar to GP|9294238|dbj| contains similarity t... 74 2e-13
BM084967 72 9e-13
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 630 bits (1625), Expect = 0.0
Identities = 345/861 (40%), Positives = 499/861 (57%), Gaps = 6/861 (0%)
Frame = +1
Query: 15 LPPVRDVEFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPWGAPVL 74
LPP R+ + I + +G GP+ + PYR + +++ ++++ +G I+PS SP+ P+L
Sbjct: 1735 LPPQREQDHAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSPFSLPIL 1914
Query: 75 LVKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVK 134
LVKKKDG R C DYR LN TVK+ +P+P +D+L+D+L G FSK+DL+S YHQI V+
Sbjct: 1915 LVKKKDGSWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQ 2094
Query: 135 TEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS* 194
ED KTAFRT +GHYE+LVMPFG+TNAPA F MN+IF L +FV+VF DDILIYS
Sbjct: 2095 PEDREKTAFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSA 2274
Query: 195 DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAW 254
K+H +HL VL L++ +L+A SKC F EV++LGH +S G+ ++ KV+ VL W
Sbjct: 2275 SWKDHLKHLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDW 2454
Query: 255 EQPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERL 314
P V ++R F+GL GYYRRFI+ +A I GPLT L +KD F W + +F LK+ +
Sbjct: 2455 PTPNNVKQLRGFLGLTGYYRRFIKSYANIAGPLTDLLQKDS-FLWNNEAEAAFVKLKKAM 2631
Query: 315 TTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELTA 374
T +PVL LP +P+ + DAS G+G VL Q+ +AY S++L + + EL A
Sbjct: 2632 TEAPVLSLPDFSQPFILETDASGIGVGAVLGQNGHPIAYFSKKLAPRMQKQSAYTRELLA 2811
Query: 375 IVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFTLQYHPGKA 434
I +L +RHYL G F I +D +SLK L DQ Q+ W+ Y+F ++Y PGK
Sbjct: 2812 ITEALSKFRHYLLGNKFIIRTDQRSLKSLMDQSLQTPEQQAWLHKFLGYDFKIEYKPGKD 2991
Query: 435 NVVADALSRKMHVSLKMVKELELLEQFRDMSLGVTPYEGMLKFGMIRIASGLMEEIRERQ 494
N ADALSR M S +EE+R R
Sbjct: 2992 NQAADALSR------------------------------MFMLAWSEPHSIFLEELRARL 3081
Query: 495 LQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNPELRRLIMDEGHKSKWRIHP 554
+ D L + QG D + +L KDRV +P E+ I+ E H S H
Sbjct: 3082 ISDPHLKQLMETYKQGADASHYTVREGLLYWKDRVVIPAEEEIVNKILQEYHSSPIGGHA 3261
Query: 555 GMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKWDSIS 614
G+T+ LK F+WP M++ V Y+ CL CQ+AK + PAG+LQ L +P+ W+ ++
Sbjct: 3262 GITRTLARLKAQFYWPKMQEDVKAYIQKCLICQQAKSNNTLPAGLLQPLPIPQQVWEDVA 3441
Query: 615 MDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHGVSTSI 674
MDF+ LP + I V++DRLTK AHF+P + YN + +AE +++ IV+LHG+ SI
Sbjct: 3442 MDFITGLPNSFGL-SVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSHIVKLHGIPRSI 3618
Query: 675 VSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSW 734
VSD D FTS FW L GT L +SSAYHPQ DGQ+E + LE LR ++ W
Sbjct: 3619 VSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYLRCFTYEHPKGW 3798
Query: 735 DDLQPLIEFTYNNNFHASIGMAPYEALYGRKCRTPLCWYQDGESLLIGPELLQQTTEK-- 792
P EF YN +H S+GM P+ ALYGR+ P + S+ E+ +Q T++
Sbjct: 3799 VKALPWAEFWYNTAYHMSLGMTPFRALYGRE---PPTLTRQACSIDDPAEVREQLTDRDA 3969
Query: 793 -VKQIREKMRASQSRQKSYADQRRRTLEFEEGDHVFLRV---TQTTGVGRAIKSRKLTPK 848
+ +++ + +Q K AD++R + F+ GD V +++ Q + V R K++KL+ +
Sbjct: 3970 LLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQPYRQHSAVLR--KNQKLSMR 4143
Query: 849 FIGPYQITRRVGPVAYQIALP 869
+ GP+++ ++G VAY++ LP
Sbjct: 4144 YFGPFKVLAKIGDVAYKLELP 4206
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 154 bits (388), Expect = 2e-37
Identities = 91/254 (35%), Positives = 136/254 (52%), Gaps = 19/254 (7%)
Frame = +1
Query: 49 LKSQLEDLSAKGFIRP-SVSPWGAPVLLVKKKDGKS------------------RLCVDY 89
++ ++ L G I P S S W +PVL+V KK+G + +LC+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 90 RQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVKTEDIPKTAFRTRYGH 149
R+LN+AT K+ +PLP +D ++++L G A + +D Y+QI V +D K AF +G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 150 YEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS*DTKEHEEHLCQVLHV 209
+ Y +PFG+ NAP F M IF + +++ + VF+DD ++ + + L VL
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 210 LREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAWEQPKTVTEIRSFVGL 269
E L N KC F + E LGH IS GI VD K++ + P V IRSF+G
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 270 AGYYRRFIEGFAKI 283
A +YRRFI+ F K+
Sbjct: 721 ARFYRRFIKDFTKV 762
>CF922488
Length = 741
Score = 150 bits (380), Expect = 2e-36
Identities = 86/246 (34%), Positives = 137/246 (54%)
Frame = +3
Query: 76 VKKKDGKSRLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVKT 135
V K+DGK +CVDYR LN A+ K+++PLP I+ L+D + FS +D S Y+QI++
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 136 EDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS*D 195
ED+ KT F T +G + Y M FG+ N A + M +F + + + + V++DD+++ S
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 196 TKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAWE 255
+EH +L ++ LR+ +L NP+KC F ++ L + S GI VD KV+ +L
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 256 QPKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERLT 315
+P T +++ F+G Y RFI PL L K+Q W C +F+ +K+ L
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 316 TSPVLI 321
VL+
Sbjct: 723 NPHVLV 740
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 144 bits (363), Expect = 2e-34
Identities = 88/254 (34%), Positives = 131/254 (50%), Gaps = 19/254 (7%)
Frame = +1
Query: 49 LKSQLEDLSAKGFIRP-SVSPWGAPVLLVKKKDGKS------------------RLCVDY 89
++ ++ L G I P S S W +PV +V KK G + R+C+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 90 RQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVKTEDIPKTAFRTRYGH 149
R+LN+AT K+ YPLP +D ++ +L + + +D S Y+QI V +D KTAF +
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 150 YEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS*DTKEHEEHLCQVLHV 209
+ Y MPFG+ NA F M IF + +++ + VF+DD + +L +VL
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 210 LREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAKVETVLAWEQPKTVTEIRSFVGL 269
+ L N KC F ++E LGH ISK GI V K++ + P V I SF+G
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 270 AGYYRRFIEGFAKI 283
G+YRRFI+ F K+
Sbjct: 721 VGFYRRFIKDFTKV 762
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 144 bits (363), Expect = 2e-34
Identities = 70/161 (43%), Positives = 103/161 (63%)
Frame = +1
Query: 258 KTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERLTTS 317
K+V +IRSF GLA +YRRF+ F+ + PL +L +K+ F W E + +F LKE+LT +
Sbjct: 106 KSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKA 285
Query: 318 PVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELTAIVF 377
PVL LP + +E+ CDAS G+ VL+Q +AY S +L + NYPT+D EL A++
Sbjct: 286 PVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYALIR 465
Query: 378 SLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWME 418
+ + W H+L F I SDH+SLKY+ + +LN R +W+E
Sbjct: 466 APQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKWVE 588
>CO982196
Length = 812
Score = 115 bits (289), Expect = 7e-26
Identities = 65/161 (40%), Positives = 91/161 (56%)
Frame = +1
Query: 526 KDRVCVPTNPELRRLIMDEGHKSKWRIHPGMTKMYQDLKLNFWWPGMKKQVVEYVAACLT 585
KDR+ + N L++ E S H G + ++ + +W GMKK +YVAAC
Sbjct: 331 KDRLVLSKNSTKIPLLLKELQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYVAACEI 510
Query: 586 CQKAKVEHQRPAGMLQSLDVPEWKWDSISMDFVVALPRTQKRHDSIWVIVDRLTKSAHFL 645
C++ K PAG+L L +P W ISMDF+ LP+ Q + D+I V+VDRLTK AHF
Sbjct: 511 CRRNKTSTLSPAGLL*LLPIPTKVWTDISMDFIGGLPKAQGK-DNILVVVDRLTKYAHFF 687
Query: 646 PGRTTYNVEKLAEIYVAEIVRLHGVSTSIVSDSDPKFTSHF 686
Y +++AE+++ E+VRLHG SIVSD F S F
Sbjct: 688 ALSHPYTAKEVAELFIKELVRLHGFPASIVSDXXRLFMSLF 810
>BI425021
Length = 426
Score = 110 bits (275), Expect = 3e-24
Identities = 57/125 (45%), Positives = 78/125 (61%)
Frame = -1
Query: 603 LDVPEWKWDSISMDFVVALPRTQKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVA 662
L VP+ W+ +SMDF+V LP H +I+V+V+R +K H T++ +A +++
Sbjct: 417 LPVPQRPWEDLSMDFIVGLP-PYHGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASLFLN 241
Query: 663 EIVRLHGVSTSIVSDSDPKFTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDL 722
+++LHG SIVSD DP F SHFW L GT LR+SSAYHPQ DGQTE + +E
Sbjct: 240 IVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVIEQY 61
Query: 723 LRACV 727
LRA V
Sbjct: 60 LRAFV 46
>BE802896
Length = 416
Score = 108 bits (270), Expect = 1e-23
Identities = 57/137 (41%), Positives = 82/137 (59%), Gaps = 4/137 (2%)
Frame = -2
Query: 265 SFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERLTTSPVLILPQ 324
SF+G AG+YRRFI F K+ PL+ L +K+ F + + CK +F LK L T+P++ P
Sbjct: 415 SFLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPD 236
Query: 325 PEEPYEVYCDASHQGLGCVLMQHW----KVVAYASRQLKTHEKNYPTHDLELTAIVFSLK 380
P+E+ CDAS+ LG VL Q +V+ Y+SR L + NY T + EL AIVF+L+
Sbjct: 235 WTAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALE 56
Query: 381 IWRHYLYGCTFTIFSDH 397
+ YL G ++ DH
Sbjct: 55 KFHSYLLGTRIIVYIDH 5
>TC211627
Length = 1034
Score = 70.1 bits (170), Expect(2) = 2e-22
Identities = 37/78 (47%), Positives = 44/78 (55%)
Frame = +2
Query: 682 FTSHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLI 741
F S W L GTKLR S+AYHPQ DGQTE + LE LRA V D+ W L
Sbjct: 5 FISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSLA 184
Query: 742 EFTYNNNFHASIGMAPYE 759
E YN + H+ IG +P+E
Sbjct: 185 E*CYNTSVHSGIGFSPFE 238
Score = 55.1 bits (131), Expect(2) = 2e-22
Identities = 30/80 (37%), Positives = 42/80 (52%)
Frame = +3
Query: 799 KMRASQSRQKSYADQRRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIGPYQITRR 858
+++ Q K AD RR L F GD V++R+ KL+ +F GPYQI R
Sbjct: 357 RLQKYQDSMKRIADSHRRDLTFNIGDWVYVRL*PYRQTSIQSTYTKLSKRFYGPYQIQAR 536
Query: 859 VGPVAYQIALPPFLPIFTMY 878
VG VAY++ LPP I ++
Sbjct: 537 VGQVAYRLQLPPTSKIHPIF 596
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 104 bits (259), Expect = 2e-22
Identities = 55/143 (38%), Positives = 86/143 (59%)
Frame = +3
Query: 84 RLCVDYRQLNKATVKNRYPLPRIDDLMDQLRGDAVFSKIDLKSSYHQIRVKTEDIPKTAF 143
R+CVDYR LN+A+ K+ +PLP ID LM + A+FS +D S Y+QI++ ED+ KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 144 RTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFLDRFVVVFIDDILIYS*DTKEHEEHL 203
T +G + Y VM FG+ N A + M +F + + + + ++D+++ S +EH +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 204 CQVLHVLREKKLYANPSKCEFWL 226
+ LR+ +L NP KC F L
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>BI317507
Length = 359
Score = 101 bits (252), Expect = 1e-21
Identities = 53/113 (46%), Positives = 73/113 (63%), Gaps = 1/113 (0%)
Frame = -1
Query: 188 DILIYS*DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISKEGIVVDPAK 247
+ILIYS D K H HL VL VL++++L AN KC F + +LGHVISK+ + +D K
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 248 VETVLAWEQPKTVTEIRSFVGLAGYYRRFIEGFAKIV-GPLTQLTRKDQPFAW 299
V++V+ W PK V + SF+ L GYYR+FI+ + K+ PLT LT+ D F W
Sbjct: 173 VKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLTKND-GFKW 18
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 97.8 bits (242), Expect = 2e-20
Identities = 48/133 (36%), Positives = 81/133 (60%)
Frame = +2
Query: 119 FSKIDLKSSYHQIRVKTEDIPKTAFRTRYGHYEYLVMPFGVTNAPAVFMDYMNRIFHEFL 178
FS +D S Y+QI + ED+ KT F T +G + Y VM FG+ N A + M +FH+ +
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 179 DRFVVVFIDDILIYS*DTKEHEEHLCQVLHVLREKKLYANPSKCEFWLEEVNFLGHVISK 238
+ + V++DD++ S EH +LC++ L++ +L NP+KC F ++ LG ++S+
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 239 EGIVVDPAKVETV 251
+GI +DP KV+ +
Sbjct: 362 KGIEIDPEKVKAL 400
>AW570005
Length = 413
Score = 93.2 bits (230), Expect = 5e-19
Identities = 51/132 (38%), Positives = 72/132 (53%)
Frame = -2
Query: 257 PKTVTEIRSFVGLAGYYRRFIEGFAKIVGPLTQLTRKDQPFAWTEACKTSFQTLKERLTT 316
P+T +R F+ L G+YRRFI+G+A + PL+ L KD F W+ +FQ LK +T
Sbjct: 406 PRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDS-FVWSPEADVAFQALKNVVTN 230
Query: 317 SPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVAYASRQLKTHEKNYPTHDLELTAIV 376
+ VL LP +P+ V DAS +G VL Q +A+ S++ T+ EL AI
Sbjct: 229 TLVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAAIT 50
Query: 377 FSLKIWRHYLYG 388
+K WR YL G
Sbjct: 49 NVVKKWRQYLLG 14
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo},
partial (9%)
Length = 410
Score = 93.2 bits (230), Expect = 5e-19
Identities = 47/64 (73%), Positives = 53/64 (82%)
Frame = -3
Query: 809 SYADQRRRTLEFEEGDHVFLRVTQTTGVGRAIKSRKLTPKFIGPYQITRRVGPVAYQIAL 868
SY D+RR+ LEFE GDHVFLRVT TGVGRA+KS KLTP FIGP+QI ++V VAYQIAL
Sbjct: 408 SYHDKRRKDLEFEVGDHVFLRVTP*TGVGRALKS*KLTPHFIGPFQILKKVDFVAYQIAL 229
Query: 869 PPFL 872
PP L
Sbjct: 228 PPSL 217
>TC234828
Length = 857
Score = 90.9 bits (224), Expect = 2e-18
Identities = 39/63 (61%), Positives = 52/63 (81%)
Frame = +3
Query: 10 EEVPRLPPVRDVEFVIDIVRGFGPISIAPYRMAPAELVELKSQLEDLSAKGFIRPSVSPW 69
++V LPP R+VEF+ID+V G P+SIAPYRM+P EL E+K+Q++DL +K F+RPS SPW
Sbjct: 666 DDVCELPPEREVEFIIDVVPGANPVSIAPYRMSPVELAEVKAQVQDLLSKQFVRPSASPW 845
Query: 70 GAP 72
GAP
Sbjct: 846 GAP 854
>BQ299538
Length = 426
Score = 60.5 bits (145), Expect(2) = 2e-17
Identities = 33/71 (46%), Positives = 40/71 (55%)
Frame = +3
Query: 695 GTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEFTYNNNFHASIG 754
GT L++S++YHP IDGQT LE LR V D E+ YN NFHAS G
Sbjct: 144 GTYLKMSTSYHP*IDGQTVN--HCLETFLRCFVADQPKM*VQWLSWAEYWYNTNFHASTG 317
Query: 755 MAPYEALYGRK 765
P+E +YGRK
Sbjct: 318 TTPFEVVYGRK 350
Score = 48.1 bits (113), Expect(2) = 2e-17
Identities = 21/40 (52%), Positives = 26/40 (64%)
Frame = +1
Query: 651 YNVEKLAEIYVAEIVRLHGVSTSIVSDSDPKFTSHFWGAL 690
Y+ LAEI+ E+V LHGV S++SD DP F S FW L
Sbjct: 13 YSARVLAEIFTKEVVHLHGVPASVLSDEDPIFVSSFWKEL 132
>BI424213
Length = 426
Score = 84.0 bits (206), Expect = 3e-16
Identities = 41/87 (47%), Positives = 51/87 (58%)
Frame = +1
Query: 684 SHFWGALHDALGTKLRLSSAYHPQIDGQTERTIQSLEDLLRACVLDNRGSWDDLQPLIEF 743
SHFW L LGTKL S+ HPQ DGQT+ +SL LLRA + N SWD+ P +EF
Sbjct: 4 SHFWKTLWAKLGTKLLFSTTCHPQTDGQTKVVNRSLSTLLRALLKGNHKSWDEYLPHVEF 183
Query: 744 TYNNNFHASIGMAPYEALYGRKCRTPL 770
YN H + +P+E +YG TPL
Sbjct: 184 AYNRGVHRTTKQSPFEVVYGFNPLTPL 264
>BQ627806
Length = 435
Score = 55.8 bits (133), Expect(2) = 9e-14
Identities = 27/68 (39%), Positives = 40/68 (58%)
Frame = +1
Query: 367 THDLELTAIVFSLKIWRHYLYGCTFTIFSDHKSLKYLFDQKELNMRQRRWMEFIKDYEFT 426
T+ EL AI ++K WR YL G F I +DH+SLK L Q Q+ ++ + +++T
Sbjct: 232 TYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQTPEQQIYLARLMGFDYT 411
Query: 427 LQYHPGKA 434
+QY GKA
Sbjct: 412 IQYRAGKA 435
Score = 40.0 bits (92), Expect(2) = 9e-14
Identities = 24/61 (39%), Positives = 32/61 (52%)
Frame = +3
Query: 293 KDQPFAWTEACKTSFQTLKERLTTSPVLILPQPEEPYEVYCDASHQGLGCVLMQHWKVVA 352
KDQ F W E +F LK L +PVL LP + V DAS G+G +L Q+ +A
Sbjct: 12 KDQ-FHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHPLA 188
Query: 353 Y 353
+
Sbjct: 189 F 191
>BI317638 weakly similar to GP|9294238|dbj| contains similarity to reverse
transcriptase~gene_id:K11J14.5 {Arabidopsis thaliana},
partial (5%)
Length = 420
Score = 74.3 bits (181), Expect = 2e-13
Identities = 40/126 (31%), Positives = 65/126 (50%)
Frame = -2
Query: 565 LNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKWDSISMDFVVALPRT 624
L+F + + + + CL CQ K E +R +L L VP W+ +S+DF+ L
Sbjct: 380 LDFQRTSIGLECAQMLPNCLDCQHTKYETKRIVDLLCPLLVPHRPWEDLSLDFITGL-LP 204
Query: 625 QKRHDSIWVIVDRLTKSAHFLPGRTTYNVEKLAEIYVAEIVRLHGVSTSIVSDSDPKFTS 684
H +I V+VD +K H +++ +A +++ + +LHG+ S+VSD D F S
Sbjct: 203 YHVHTAILVVVDHFSKGIHLGMLPSSHTAHTVACLFIDSVAKLHGLPRSLVSDCDLLFVS 24
Query: 685 HFWGAL 690
HFW L
Sbjct: 23 HFWQEL 6
>BM084967
Length = 426
Score = 72.4 bits (176), Expect = 9e-13
Identities = 48/144 (33%), Positives = 72/144 (49%)
Frame = -2
Query: 494 QLQDVFLLEKRNLVIQGKDPEFKIGTDNILRCKDRVCVPTNPELRRLIMDEGHKSKWRIH 553
QLQ L E P+ + D IL+ + +P+ ++ E H S H
Sbjct: 413 QLQQAILAEPTTY------PDHSVIQDLILK-NGCIWLPSGFSFIPTLLLEYHSSPTDAH 255
Query: 554 PGMTKMYQDLKLNFWWPGMKKQVVEYVAACLTCQKAKVEHQRPAGMLQSLDVPEWKWDSI 613
G+TK L NF W G++K V ++VAACL CQ K E Q+ AG+L L VP W+ +
Sbjct: 254 IGVTKTMARLSENFTWIGIRKDVEQFVAACLDCQYTKYEAQKMAGLLCPLPVPCRPWEDL 75
Query: 614 SMDFVVALPRTQKRHDSIWVIVDR 637
S +F++ L + + +I V+V R
Sbjct: 74 SFNFIIGLSEF-RGYTAILVVVGR 6
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.328 0.141 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,478,519
Number of Sequences: 63676
Number of extensions: 609921
Number of successful extensions: 3515
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 3470
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3501
length of query: 968
length of database: 12,639,632
effective HSP length: 106
effective length of query: 862
effective length of database: 5,889,976
effective search space: 5077159312
effective search space used: 5077159312
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0306.4


