

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0306.1
(1224 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
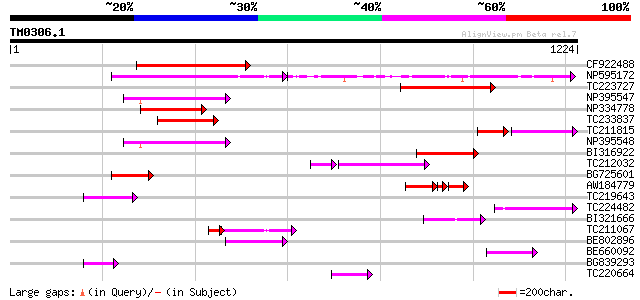
Score E
Sequences producing significant alignments: (bits) Value
CF922488 214 2e-55
NP595172 polyprotein [Glycine max] 192 8e-49
TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 188 1e-47
NP395547 reverse transcriptase [Glycine max] 158 2e-38
NP334778 reverse transcriptase [Glycine max] 153 5e-37
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 142 9e-34
TC211815 92 1e-33
NP395548 reverse transcriptase [Glycine max] 138 1e-32
BI316922 126 7e-29
TC212032 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 92 2e-24
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 109 6e-24
AW184779 72 3e-23
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 94 4e-19
TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 91 4e-18
BI321666 83 8e-16
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 61 4e-13
BE802896 66 8e-11
BE660092 weakly similar to GP|9884624|dbj retroelement pol polyp... 60 4e-09
BG839293 60 6e-09
TC220664 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial... 58 2e-08
>CF922488
Length = 741
Score = 214 bits (544), Expect = 2e-55
Identities = 108/245 (44%), Positives = 153/245 (62%)
Frame = +3
Query: 275 VKKSNGKWRMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYA 334
V K +GK MC DY DLN PKD +PLP+I+ LVD + F S MD +SGY+QI++
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 335 PDEEKTAFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEE 394
D EKT F+T +CY+ M FGLKN GATYQR M +F + + +E+Y+DDM+VKS
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 395 MGGHCLDLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQ 454
H ++L + F +RK+ +RLNP KC F ++S K L F+ + RGIEV+ +K K ILEM
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 455 SPTSVKEVQKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLS 514
P + K+VQ +GR+ + RF+ + P F L KN+ +W +C AF+ +K+ L
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 515 KPPIL 519
P +L
Sbjct: 723 NPHVL 737
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 192 bits (488), Expect = 8e-49
Identities = 116/379 (30%), Positives = 190/379 (49%)
Frame = +1
Query: 221 HKLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNG 280
H + L G P+ + + ++ +++ G I+ P L +++VKK +G
Sbjct: 1759 HAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSPFSLP-ILLVKKKDG 1935
Query: 281 KWRMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKT 340
WR CTDY LN KDS+P+P +D+L+D G S +D SGYHQI + D EKT
Sbjct: 1936 SWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKT 2115
Query: 341 AFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCL 400
AF T+ +Y + MPFGL NA AT+Q LM+++F+ + + + ++ DD+++ S H
Sbjct: 2116 AFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLK 2295
Query: 401 DLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVK 460
L +++H + KCSFG +LG ++ G+ + K +A+L+ +P +VK
Sbjct: 2296 HLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVK 2475
Query: 461 EVQKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILS 520
+++ +G RF+ + A P L+K+ F W +E E AF LK+ +++ P+LS
Sbjct: 2476 QLRGFLGLTGYYRRFIKSYANIAGPLTDLLQKDS-FLWNNEAEAAFVKLKKAMTEAPVLS 2652
Query: 521 RPIPGTPLSVFISISDNAVSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAALALII 580
P P + S V +VL Q I YF S L + + LA+
Sbjct: 2653 LPDFSQPFILETDASGIGVGAVLGQNGHP---IAYF-SKKLAPRMQKQSAYTRELLAITE 2820
Query: 581 SARKLRPYFQGFQIKVKTD 599
+ K R Y G + ++TD
Sbjct: 2821 ALSKFRHYLLGNKFIIRTD 2877
Score = 119 bits (298), Expect = 8e-27
Identities = 159/648 (24%), Positives = 263/648 (40%), Gaps = 27/648 (4%)
Frame = +1
Query: 601 PLRQVLQKPDLAGRMVSWAVELSEFGIVFEKKGQVKAQVLA--DFVNEMSPEVKVSEEAE 658
PL +LQK W E +E V KK +A VL+ DF
Sbjct: 2548 PLTDLLQKDSFL-----WNNE-AEAAFVKLKKAMTEAPVLSLPDF------------SQP 2673
Query: 659 WILSVDGSSYLKGSGAGVVLEGPGGVIIEQSLKFDFKASNNQAEYEAIIAGINLAIEMNV 718
+IL D S G G G VL G I S K + Q+ Y + I A+
Sbjct: 2674 FILETDAS----GIGVGAVLGQNGHPIAYFSKKLAPRMQK-QSAYTRELLAITEALSKFR 2838
Query: 719 HCL-----VIKTDSQLVANQIKGDYQAKDIQ--LAKYLTKTQELMKRMDSVQVNHVPREE 771
H L +I+TD + + + + Q + Q L K+L ++ + P ++
Sbjct: 2839 HYLLGNKFIIRTDQRSLKSLMDQSLQTPEQQAWLHKFLGY---------DFKIEYKPGKD 2991
Query: 772 NTRADVLCKL--ASTKKPGNNKSVIQETLKSPSINEDDVVMVTGAAPSDWMDRIKMCLEA 829
N AD L ++ + +P S+ E L++ I++ + + M+ K
Sbjct: 2992 NQAADALSRMFMLAWSEP---HSIFLEELRARLISDPHLKQL--------METYKQ---- 3126
Query: 830 DGADLALFSKDQVREASHYVLLGDQLYRRG-VGVPLLRCVTRDEADRIMFEVHEGVCASH 888
GAD ASHY + LY + V +P + ++I+ E H H
Sbjct: 3127 -GAD-----------ASHYTVREGLLYWKDRVVIP----AEEEIVNKILQEYHSSPIGGH 3258
Query: 889 VGGRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKCQIYADLHRAPPEVLSSMSSAWPFAMW 948
G A+ L+A FYWP ++ D Y +KC CQ + P +L + P +W
Sbjct: 3259 AGITRTLAR-LKAQFYWPKMQEDVKAYIQKCLICQQAKSNNTLPAGLLQPLPI--PQQVW 3429
Query: 949 GVDILGPFT---PAGAQIRFVLVAVDYFTKW-----IEAESMAKITAEKVKKFYWRKIIC 1000
D+ F P + ++V +D TK+ ++A+ +K+ AE + I+
Sbjct: 3430 E-DVAMDFITGLPNSFGLSVIMVVIDRLTKYAHFIPLKADYNSKVVAEA----FMSHIVK 3594
Query: 1001 RFGVPATLVSDNGTQFTSRIVRDFCNEMGIEMRFASVEHPQSNGQVEAANKVILNGIKKR 1060
G+P ++VSD FTS + G + +S HPQS+GQ E NK + ++
Sbjct: 3595 LHGIPRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYLRCF 3774
Query: 1061 LGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLTYGVDAMVPVEIQDMTFRVAAYDENE 1120
+ W L + YNT + G TPFR YG E +T + + D+
Sbjct: 3775 TYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYG------REPPTLTRQACSIDDPA 3936
Query: 1121 NHENRLIDLNLAEEVKTEVRLRQAAVKQRSERRYNTRVVPRHMQVGDLVL-------RRK 1173
+L D + + ++++ +Q +R+ + + + Q+GD VL +
Sbjct: 3937 EVREQLTD---RDALLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQPYRQHS 4107
Query: 1174 AKGPDDSKLSPNWEGPYRILRDLGQGAYHLEELSGRRIPRAWNAQHLR 1221
A + KLS + GP+++L +G AY LE S RI ++ L+
Sbjct: 4108 AVLRKNQKLSMRYFGPFKVLAKIGDVAYKLELPSAARIHPVFHVSQLK 4251
>TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (9%)
Length = 843
Score = 188 bits (478), Expect = 1e-47
Identities = 88/208 (42%), Positives = 132/208 (63%), Gaps = 1/208 (0%)
Frame = +1
Query: 843 REASHYVLLGDQLYRRGVGVPLLRCVTRDEADRIMFEVHEGVCASHVGGRSLAAKVLRAG 902
R A+ + + G LY+R + LRCV EA+ ++ EVHEG +H G ++A K+LRAG
Sbjct: 220 RLAAGFFMSGSILYKRNHDMKPLRCVDAREANHMIEEVHEGSFGTHANGHAMARKILRAG 399
Query: 903 FYWPTLKNDCMGYAKKCEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDILGPFTP-AGA 961
+YW T+++DC + +KC KCQ +AD APP L+ MSS WPF+MWG+D++G P A
Sbjct: 400 YYWLTMESDCCVHVRKCHKCQAFADNVNAPPHPLNVMSSPWPFSMWGIDVIGAIEPKASN 579
Query: 962 QIRFVLVAVDYFTKWIEAESMAKITAEKVKKFYWRKIICRFGVPATLVSDNGTQFTSRIV 1021
RF+LVA+DYFTKW+EA S + V +F ++IICR+G+P +++DNGT ++++
Sbjct: 580 GHRFILVAIDYFTKWVEAASYTDVMRGVVVRFIKKEIICRYGLPRKIITDNGTNLNNKMM 759
Query: 1022 RDFCNEMGIEMRFASVEHPQSNGQVEAA 1049
+ C E I+ + P+ N VE A
Sbjct: 760 GEICEEFKIQHHNPTPYRPKMN*AVEVA 843
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 158 bits (399), Expect = 2e-38
Identities = 86/248 (34%), Positives = 130/248 (51%), Gaps = 18/248 (7%)
Frame = +1
Query: 247 VKAETNKLIDAGFIREVKYPTWLANVVMVKKSNG------------------KWRMCTDY 288
V+ E KL++AG I + +W++ V +V K G +WRMC DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 289 TDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQAN 348
LN+ KD YPLP +D+++ R + +D YSGY+QI + D+EKTAF +
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 349 YCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGE 408
+ Y+ MPFGL NA T+QR M +F+ V + +E+++DD G +L +
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 409 IRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKEVQKLIGR 468
K N+ LN EKC F +Q G LG I++RGIEV +K I ++ P +VK + +G
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 469 IAALSRFL 476
+ RF+
Sbjct: 721 VGFYRRFI 744
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 153 bits (386), Expect = 5e-37
Identities = 69/143 (48%), Positives = 96/143 (66%)
Frame = +3
Query: 283 RMCTDYTDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAF 342
RMC DY DLN+ PKD++PLP+ID L+ + F + S MD +SGY+QI+M D EKT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 343 MTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDL 402
+T +CY+ M FGLKN GATY R M +F+ + + +E YVD+M+ KS H ++L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 403 AEAFGEIRKHNMRLNPEKCSFGI 425
FG++RK+ +RLNP KC FG+
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 142 bits (358), Expect = 9e-34
Identities = 63/132 (47%), Positives = 94/132 (70%)
Frame = +2
Query: 319 SLMDAYSGYHQIRMYAPDEEKTAFMTNQANYCYQTMPFGLKNAGATYQRLMDRVFEGQVG 378
S MD +SGY+QI M D EKT F+T + Y+ M FGLKN GATYQR M +F +
Sbjct: 5 SFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMMH 184
Query: 379 RNMEIYVDDMVVKSEEMGGHCLDLAEAFGEIRKHNMRLNPEKCSFGIQSGKFLGFMITRR 438
+ +E+YVDDM+ KS H ++L + FG ++K+ ++LNP KC+FG++SGK LGF+++++
Sbjct: 185 KEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQK 364
Query: 439 GIEVNPDKCKAI 450
GIE++P+K KA+
Sbjct: 365 GIEIDPEKVKAL 400
>TC211815
Length = 704
Score = 91.7 bits (226), Expect(2) = 1e-33
Identities = 54/144 (37%), Positives = 83/144 (57%), Gaps = 2/144 (1%)
Frame = +2
Query: 1083 PQSTTGETPFRLTYGVDAMVPVEIQDMTFRVAAYDENENHENRLIDLNLAEEVKTEVRLR 1142
PQSTT ETPF L YG+ M+P+E+ ++ + E N E IDL+L ++V+ + +
Sbjct: 227 PQSTTHETPF*LIYGISVMLPIEVGEVFL*RHYFAEV*NKEALQIDLDLIKQVREDTVIM 406
Query: 1143 QAAVKQRSERRYNTRVVPRHMQVGDLV--LRRKAKGPDDSKLSPNWEGPYRILRDLGQGA 1200
A KQR R +N+++ G + ++R + SK + N EGP++I + GA
Sbjct: 407 T*AFKQRMTRCFNSKLPSTV*GRGPSMEGIQRSLEVLVRSKFTTN*EGPFKIRHNSKNGA 586
Query: 1201 YHLEELSGRRIPRAWNAQHLRYYY 1224
Y LEELSG+ + R WN+ HL+ YY
Sbjct: 587 YKLEELSGKVVLRIWNSMHLKVYY 658
Score = 71.6 bits (174), Expect(2) = 1e-33
Identities = 32/67 (47%), Positives = 47/67 (69%)
Frame = +3
Query: 1011 DNGTQFTSRIVRDFCNEMGIEMRFASVEHPQSNGQVEAANKVILNGIKKRLGDAKGLWAD 1070
DNG QFT+R + +F + + I+ R SV+HPQ+N + EAANKVIL +KK L A G W +
Sbjct: 3 DNGLQFTNRKLNEFPSGLNIKHRVTSVKHPQTNRRAEAANKVILGDLKKLLDGANGRWVE 182
Query: 1071 ELLTVVW 1077
+L+ ++W
Sbjct: 183 DLVEILW 203
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 138 bits (348), Expect = 1e-32
Identities = 78/248 (31%), Positives = 127/248 (50%), Gaps = 18/248 (7%)
Frame = +1
Query: 247 VKAETNKLIDAGFIREVKYPTWLANVVMVKKSNG------------------KWRMCTDY 288
V+ E KL++ G I + W++ V++V K G W++C DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 289 TDLNKHCPKDSYPLPNIDKLVDRASGFGMLSLMDAYSGYHQIRMYAPDEEKTAFMTNQAN 348
LN+ KD +PLP +D++++R +G +DAY GY+QI + D+EK AF
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 349 YCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEIYVDDMVVKSEEMGGHCLDLAEAFGE 408
+ Y+ +PFGL NA T+Q M +F V +++E+++DD V + L
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 409 IRKHNMRLNPEKCSFGIQSGKFLGFMITRRGIEVNPDKCKAILEMQSPTSVKEVQKLIGR 468
+ N+ LN EKC F ++ G LG I+ RGIEV+ K I ++ P++VK ++ +G+
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 469 IAALSRFL 476
RF+
Sbjct: 721 ARFYRRFI 744
>BI316922
Length = 405
Score = 126 bits (316), Expect = 7e-29
Identities = 58/134 (43%), Positives = 86/134 (63%)
Frame = +3
Query: 879 EVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMGYAKKCEKCQIYADLHRAPPEVLSS 938
E+H G+C H + + +VLR G+Y T++ C Y KKCE+C + ++ E L +
Sbjct: 3 EMHRGICGMHSKSQLMTTRVLRVGYY**TMRKYCTEYVKKCEEC*KFGNISHLLVEELHN 182
Query: 939 MSSAWPFAMWGVDILGPFTPAGAQIRFVLVAVDYFTKWIEAESMAKITAEKVKKFYWRKI 998
+ + WPFA+ GVDIL PF + Q++++LV +D FTKWIE E +A I+ V+KF R I
Sbjct: 183 IVAPWPFAI*GVDILRPFPLSKRQVKYLLVGIDQFTKWIETEHIAIISIANVRKFV*RNI 362
Query: 999 ICRFGVPATLVSDN 1012
+C FG+P TL+SDN
Sbjct: 363 VC*FGIPNTLISDN 404
>TC212032 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (3%)
Length = 803
Score = 92.0 bits (227), Expect(2) = 2e-24
Identities = 63/201 (31%), Positives = 97/201 (47%), Gaps = 5/201 (2%)
Frame = +2
Query: 710 INLAIEMNVHCLVIKTDSQLVANQIKGDYQAKDIQLAKYLTKTQELMKRMDSVQVNHVPR 769
+ AI+ NV L + DS LV +Q++G+ + +D L Y +EL D + +HV
Sbjct: 206 VQAAIDSNVKLLKVYGDSALVIHQLRGECETRDPNLIPYQAYIKELAGFFDEISFHHVA* 385
Query: 770 EENTRADVLCKLASTKK--PGNNKSVIQETLKSPSINEDDVVMVTGAAPSDWMDRIKMCL 827
EEN AD L L S + P + I+ + + V P W IK +
Sbjct: 386 EENQMADALATLVSMFQLTPHGDLPYIEFRCRGRPAHCCLVEEERDGKP--WYFDIKRYV 559
Query: 828 EADGADLALFSKDQVRE---ASHYVLLGDQLYRRGVGVPLLRCVTRDEADRIMFEVHEGV 884
E+ L D+ R+ A+ + + G LY+R + LL CV E + ++ EVHEG
Sbjct: 560 ESKEYPLEASDNDKRRKRRLAAGFFMSGSILYKRNHDMVLLHCVNGKEVENMLGEVHEGS 739
Query: 885 CASHVGGRSLAAKVLRAGFYW 905
+H G ++A K+LRAG+YW
Sbjct: 740 FGTHSNGHAMARKILRAGYYW 802
Score = 40.0 bits (92), Expect(2) = 2e-24
Identities = 21/57 (36%), Positives = 31/57 (53%), Gaps = 1/57 (1%)
Frame = +1
Query: 650 EVKVSEEAE-WILSVDGSSYLKGSGAGVVLEGPGGVIIEQSLKFDFKASNNQAEYEA 705
E K+ E+ + WI+ D +S + G G G +L P I + + F +NN AEYEA
Sbjct: 22 EEKLDEDRDKWIVWFDRASNVLGHGVGAILVSPDNQCIPFTTRLGFDCTNNMAEYEA 192
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 109 bits (273), Expect = 6e-24
Identities = 54/90 (60%), Positives = 64/90 (71%)
Frame = -3
Query: 221 HKLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVMVKKSNG 280
HKLA+ VK + Q KRK+ EE+ Q V E KL A FIR++ Y T L +VVMVKK NG
Sbjct: 277 HKLAICNDVKLVTQRKRKIREERCQTV*QEVVKLAIASFIRDINYST*LFSVVMVKKPNG 98
Query: 281 KWRMCTDYTDLNKHCPKDSYPLPNIDKLVD 310
KWR+CTDY DLN CPKD+YPLPNID + D
Sbjct: 97 KWRICTDYIDLN*ACPKDAYPLPNIDHMTD 8
>AW184779
Length = 432
Score = 72.4 bits (176), Expect(3) = 3e-23
Identities = 31/69 (44%), Positives = 46/69 (65%)
Frame = +1
Query: 855 LYRRGVGVPLLRCVTRDEADRIMFEVHEGVCASHVGGRSLAAKVLRAGFYWPTLKNDCMG 914
LY+R + LLRCV EA++++ EVHEG +H ++A K+LR G+YW T+++DC
Sbjct: 16 LYKRNHDMVLLRCVDAREAEQMLVEVHEGSFGTHANIHAMAQKILRVGYYWLTMESDCCI 195
Query: 915 YAKKCEKCQ 923
+ KC KCQ
Sbjct: 196 HVWKCHKCQ 222
Score = 51.6 bits (122), Expect(3) = 3e-23
Identities = 22/45 (48%), Positives = 30/45 (65%), Gaps = 1/45 (2%)
Frame = +3
Query: 947 MWGVDILGPFTPAGAQ-IRFVLVAVDYFTKWIEAESMAKITAEKV 990
MWG+D++G P + F+LVA+DYFTKW+EA S A +T V
Sbjct: 291 MWGIDVIGAIEPKASNGHHFILVAIDYFTKWVEAVSYASVTRSVV 425
Score = 24.3 bits (51), Expect(3) = 3e-23
Identities = 9/23 (39%), Positives = 15/23 (65%)
Frame = +2
Query: 923 QIYADLHRAPPEVLSSMSSAWPF 945
Q +AD APP L+ +++ WP+
Sbjct: 224 QTFADNVNAPPIPLNVLAAPWPY 292
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein (Fragment),
partial (8%)
Length = 1320
Score = 94.0 bits (232), Expect = 4e-19
Identities = 47/121 (38%), Positives = 72/121 (58%), Gaps = 5/121 (4%)
Frame = +3
Query: 160 EHKMTP-EEETKTVKVG----ERNLKVGVNLTAIQEARLTQLLAENMDLFAWSAQDLPGI 214
+ +M P +EET+ V +G +R +K+G +TA L LL + D+FAWS QD+PG+
Sbjct: 954 DQEMGPHQEETELVDLGSGSGKREVKIGTGITAPIREELIILLRDYQDIFAWSYQDMPGL 1133
Query: 215 DPNFICHKLALNPGVKPIAQMKRKMGEEKAQAVKAETNKLIDAGFIREVKYPTWLANVVM 274
+ + H+L LNP P+ Q R+M E + +K E K DAGF+ +YP W+AN+V
Sbjct: 1134 SSDIVQHRLPLNPECSPVKQKLRRMKPETSLKIKEEVKK*FDAGFLAVARYPKWVANIVP 1313
Query: 275 V 275
+
Sbjct: 1314 I 1316
>TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 669
Score = 90.5 bits (223), Expect = 4e-18
Identities = 58/183 (31%), Positives = 93/183 (50%), Gaps = 5/183 (2%)
Frame = +1
Query: 1047 EAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTGETPFRLTYGVDAMVPVEI 1106
EAANK I I+K K W + L + Y T+ +++TG TPF L YG++A++P E+
Sbjct: 1 EAANKNIKKIIQKMTVSYKD-WHEMLPFALHGYRTSVRTSTGATPFSLVYGMEAVLPFEV 177
Query: 1107 QDMTFRVAA---YDENENHENRLIDLNLAEEVKTEVRLRQAAVKQRSERRYNTRVVPRHM 1163
+ + R+ A E+E + R LNL E + +QR + ++ +V R
Sbjct: 178 EVPSLRILAESGLKESEWAQTRYDQLNLIEGKRLTAMSHGRLYQQRMKSAFDKKVCLRKF 357
Query: 1164 QVGDLVLRRKAKGPDD--SKLSPNWEGPYRILRDLGQGAYHLEELSGRRIPRAWNAQHLR 1221
GDLVL++ + D K +PN+EGP+ + R GA L + G +P N+ ++
Sbjct: 358 HEGDLVLKKMSHAVKDHRGKWAPNYEGPFVVKRAFSGGALVLTNMDGEELPSPMNSDVVK 537
Query: 1222 YYY 1224
YY
Sbjct: 538 RYY 546
>BI321666
Length = 430
Score = 82.8 bits (203), Expect = 8e-16
Identities = 43/133 (32%), Positives = 72/133 (53%)
Frame = +2
Query: 894 LAAKVLRAGFYWPTLKNDCMGYAKKCEKCQIYADLHRAPPEVLSSMSSAWPFAMWGVDIL 953
++ VL++ FY P++ D +A+ C KCQ + + L ++ F WG+D +
Sbjct: 2 ISTNVLQSRFYLPSIFKDAYVHAQSCNKCQRTRSVSKRNELPLHTILEVEIFDYWGIDFV 181
Query: 954 GPFTPAGAQIRFVLVAVDYFTKWIEAESMAKITAEKVKKFYWRKIICRFGVPATLVSDNG 1013
GPF P+ + ++LV VDY +KW+EA + K A+ V KF ++I R GVP L+ + G
Sbjct: 182 GPFPPSFSN-EYILVVVDYVSKWVEAVACQKSDAKIVIKFLKKQIFSRLGVPWVLIDNGG 358
Query: 1014 TQFTSRIVRDFCN 1026
+ + + F N
Sbjct: 359 SHLCNA*LARFFN 397
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 60.8 bits (146), Expect(2) = 4e-13
Identities = 48/162 (29%), Positives = 74/162 (45%), Gaps = 1/162 (0%)
Frame = +1
Query: 458 SVKEVQKLIGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPP 517
SV +++ G + RF+P + A+P + ++KN F W ++ EQAF LKE L+K P
Sbjct: 109 SVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKAP 288
Query: 518 ILSRPIPGTPLSVFISISDNAVSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAALA 577
+L+ P + S V +VLLQ I YF S L A L Y +K A
Sbjct: 289 VLALPDFSKTFELECDASGVGVRAVLLQGGHP---IAYF-SEKLHSATLNYPTYDKELYA 456
Query: 578 LIISARKLRPYFQGFQIKVKTDF-PLRQVLQKPDLAGRMVSW 618
LI + + + + + +D L+ + K L R W
Sbjct: 457 LIRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKW 582
Score = 33.1 bits (74), Expect(2) = 4e-13
Identities = 14/33 (42%), Positives = 22/33 (66%)
Frame = +2
Query: 430 FLGFMITRRGIEVNPDKCKAILEMQSPTSVKEV 462
F GF++ R G++++P+K KAI E P V E+
Sbjct: 23 FSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEI 121
>BE802896
Length = 416
Score = 66.2 bits (160), Expect = 8e-11
Identities = 41/134 (30%), Positives = 59/134 (43%)
Frame = -2
Query: 466 IGRIAALSRFLPCSGSKATPFFQCLRKNRVFQWTDECEQAFQSLKELLSKPPILSRPIPG 525
+G RF+ A P L+K F + D+C+ AF LK L PI+ P
Sbjct: 409 LGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPDWT 230
Query: 526 TPLSVFISISDNAVSSVLLQECKDELRIIYFVSHALQGAELRYQKIEKAALALIISARKL 585
P + S+ A+ VL Q+ R+IY+ S L A+ Y EK LA++ + K
Sbjct: 229 APFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALEKF 50
Query: 586 RPYFQGFQIKVKTD 599
Y G +I V D
Sbjct: 49 HSYLLGTRIIVYID 8
>BE660092 weakly similar to GP|9884624|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (13%)
Length = 378
Score = 60.5 bits (145), Expect = 4e-09
Identities = 30/113 (26%), Positives = 62/113 (54%), Gaps = 3/113 (2%)
Frame = -3
Query: 1029 GIEMRFASVEHPQSNGQVEAANKVILNGIKKRLGDAKGLWADELLTVVWAYNTTPQSTTG 1088
G+ R ++ HPQ+NGQ E +N+ I ++K + ++ W+ L +WA+ T ++ G
Sbjct: 352 GVVHRVSTPYHPQTNGQAEISNREIKRILEKIVQPSRKDWSTRLDDALWAHRTAYKAPIG 173
Query: 1089 ETPFRLTYGVDAMVPVEIQDMTF---RVAAYDENENHENRLIDLNLAEEVKTE 1138
+P+R+ +G +PVEI+ + + + ++ E R + L+ +E++ E
Sbjct: 172 MSPYRVVFGKACHLPVEIEHKAYWAVKTCNFSMDQAGEERKLQLSELDEIRLE 14
>BG839293
Length = 781
Score = 60.1 bits (144), Expect = 6e-09
Identities = 31/80 (38%), Positives = 48/80 (59%), Gaps = 5/80 (6%)
Frame = +1
Query: 160 EHKMTP-EEETKTVKVG----ERNLKVGVNLTAIQEARLTQLLAENMDLFAWSAQDLPGI 214
+ +M P +EET+ V +G +R +K+G +TA L LL + D+FAWS QD+PG+
Sbjct: 490 DQEMGPHQEETELVDLGIGSGKREVKIGTGITAPIREELIILLKDYQDIFAWSYQDMPGL 669
Query: 215 DPNFICHKLALNPGVKPIAQ 234
+ + H+L LNP P+ Q
Sbjct: 670 SSDIVQHQLPLNPECSPVKQ 729
>TC220664 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial (31%)
Length = 724
Score = 58.2 bits (139), Expect = 2e-08
Identities = 32/87 (36%), Positives = 49/87 (55%)
Frame = +1
Query: 696 ASNNQAEYEAIIAGINLAIEMNVHCLVIKTDSQLVANQIKGDYQAKDIQLAKYLTKTQEL 755
A+NN AEY A+I G+ A++ + I+ DS+LV QI G ++ K+ L+ +EL
Sbjct: 61 ATNNAAEYRAMILGMKYALKKGFTGIRIQGDSKLVCMQIDGSWKVKNENLSTLYNVAKEL 240
Query: 756 MKRMDSVQVNHVPREENTRADVLCKLA 782
+ S Q++HV R N+ AD LA
Sbjct: 241 KDKFSSFQISHVLRNFNSDADAQANLA 321
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,950,443
Number of Sequences: 63676
Number of extensions: 630191
Number of successful extensions: 2724
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 2694
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2711
length of query: 1224
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1116
effective length of database: 5,762,624
effective search space: 6431088384
effective search space used: 6431088384
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0306.1


