

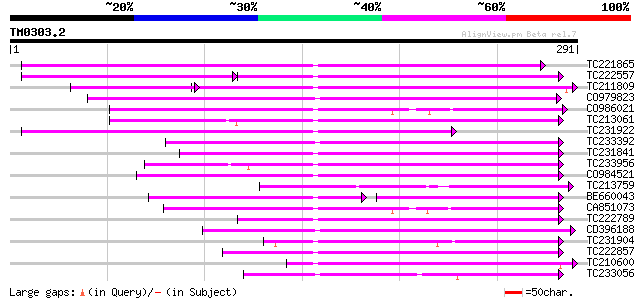
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0303.2
(291 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221865 155 2e-38
TC222557 89 1e-33
TC211809 108 1e-32
CO979823 135 3e-32
CO986021 127 8e-30
TC213061 125 3e-29
TC231922 116 1e-26
TC233392 113 9e-26
TC231841 107 6e-24
TC233956 105 2e-23
CO984521 100 6e-22
TC213759 99 2e-21
BE660043 68 4e-21
CA851073 98 5e-21
TC222789 93 2e-19
CD396188 weakly similar to GP|6175788|gb|A ORF144 {Xestia c-nigr... 90 1e-18
TC231904 similar to GB|AAL25568.1|16648753|AY058152 AT5g16150/T2... 88 5e-18
TC222857 87 7e-18
TC210600 similar to UP|Q51534 (Q51534) PilW protein, partial (7%) 83 1e-16
TC233056 82 2e-16
>TC221865
Length = 1045
Score = 155 bits (393), Expect = 2e-38
Identities = 90/272 (33%), Positives = 129/272 (47%), Gaps = 3/272 (1%)
Frame = -3
Query: 7 GDVTDFWHDDWLGTG-CLRGAFYRLYNLSRQKWNCVNTLGSWEHGRWHWKFLWRRPLVGR 65
GD FW D +G G L + R+Y +S Q+ + +GS W WKF WRRPL
Sbjct: 914 GDKFRFWEDKLVGEGETLMRKYPRMYQISCQQQQLIQQVGSHTETTWEWKFQWRRPLFDN 735
Query: 66 ELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAYHFLQ-EPTLPDTDEALVSI 124
E++ G L+ + + + + D WVW G YS SAYH +Q E +TD A I
Sbjct: 734 EVDTTIGFLEDISRFPIHRHLTDCWVWKSEPNGHYSTRSAYHLMQHEAAKANTDPAFEEI 555
Query: 125 WHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTF 184
W P+ FA R++ DR ++ NL +RQV L C C +Y E HL FSCT
Sbjct: 554 WKLKIPAKAAIFA*RLVKDRLPTKNNLRRRQV--QLNGTLCLFCRNYEEEASHLFFSCTK 381
Query: 185 SLSVWQGCLHWLGCSFIQPSSARDHLSHFRFGINKLQQRLALSI-WLAVIWSVWLARNEV 243
+ VW L W+ S ++ R H FG+ L+Q W+A+ +++W RN
Sbjct: 380 TQPVWWESLSWINTSGAFSANPRHHFLQHSFGLLGLKQYNR*KCWWVALTFTIWQHRNRT 201
Query: 244 VFRGGAANIYSVLELVKRRSWQWNKANMKGFS 275
+F N ++E W W + KGF+
Sbjct: 200 IFSNEPFNGSKMMEDAMFLVWSWFRELEKGFT 105
>TC222557
Length = 1002
Score = 89.4 bits (220), Expect(2) = 1e-33
Identities = 51/168 (30%), Positives = 76/168 (44%), Gaps = 1/168 (0%)
Frame = +1
Query: 118 DEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFH 177
DEAL +W P FAWR++ +R ++ NL +R+V L + C CN E H
Sbjct: 421 DEALEDLWQLKIPLKATTFAWRLIKERIPTKGNLWRRRV--QLNNLMCPFCNRQEEEASH 594
Query: 178 LLFSCTFSLSVWQGCLHWL-GCSFIQPSSARDHLSHFRFGINKLQQRLALSIWLAVIWSV 236
L F+C L +W L W P ++++ H K Q W+A+ WS+
Sbjct: 595 LFFNCPRILPLWWESLAWTKTVGVFSPIPRQNYMQHTLISKGKHLQARWKCWWVALTWSI 774
Query: 237 WLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSN 284
W RN +VF A N +++ W W + KGF+ + W SN
Sbjct: 775 WRHRNRIVFLNKAFNGSKLMDDAIFLVWSWFRQLEKGFAQPYNYWSSN 918
Score = 71.6 bits (174), Expect(2) = 1e-33
Identities = 39/112 (34%), Positives = 50/112 (43%), Gaps = 1/112 (0%)
Frame = +3
Query: 7 GDVTDFWHDDWL-GTGCLRGAFYRLYNLSRQKWNCVNTLGSWEHGRWHWKFLWRRPLVGR 65
GD FW D WL LR + RLYN+S Q+ + +G W WK WRR L
Sbjct: 84 GDKIRFWEDCWLMEQEPLRAKYPRLYNISCQQQKLILVMGFHSANVWEWKLEWRRHLFDN 263
Query: 66 ELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAYHFLQEPTLPDT 117
E++ LD + + + D WVW P G +S SAY L E T
Sbjct: 264 EVQAAASFLDDISWGHIDRWTLDCWVWKPEPNGQFSTRSAYRMLLEGAADQT 419
>TC211809
Length = 895
Score = 108 bits (269), Expect(2) = 1e-32
Identities = 68/209 (32%), Positives = 96/209 (45%), Gaps = 11/209 (5%)
Frame = +2
Query: 94 PSNVGIYSVNSAYHFLQEPTLP-DTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLL 152
P+ G YS SAY L E T D V +W+ PS FAWR++ DR + NL
Sbjct: 188 PTLFGNYSTKSAYRLLMEATGEFPVDRTYVDLWNLKIPSKAIVFAWRLIKDRLPTWTNLR 367
Query: 153 KRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDH-LS 211
RQV L D++C LCN E HL F CT + +W W+ P + +DH L
Sbjct: 368 VRQV--ELNDSRCPLCNSSEEDAAHLFFHCTKTEXLWWETQSWVNSLGAFPHNPKDHFLQ 541
Query: 212 HFRFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANM 271
H + R +W+A+ W +W RN VVF + + V+E +W W K
Sbjct: 542 HDNRSAGGVXARRWKCLWVALTWVIWKHRNGVVFHNHSFDGSKVMEDAILLTWSWLKVME 721
Query: 272 KGFSYSSSDWFSN---------PLLCILG 291
KGF+ S W S+ ++C++G
Sbjct: 722 KGFTGPFSFWSSHLKAAF*F*GVIICVIG 808
Score = 49.3 bits (116), Expect(2) = 1e-32
Identities = 22/66 (33%), Positives = 34/66 (51%)
Frame = +1
Query: 32 NLSRQKWNCVNTLGSWEHGRWHWKFLWRRPLVGRELEWEKGLLDALGTVQLTKGVPDAWV 91
++S Q+ ++++GS E W F WRRPL E+ L+ L V + + PD WV
Sbjct: 1 HISNQQHQSISSMGSQEDTGWE*NFSWRRPLFDNEIGMAAEFLEELAQVTIHQDRPDVWV 180
Query: 92 WMPSNV 97
W +V
Sbjct: 181WKADSV 198
>CO979823
Length = 853
Score = 135 bits (339), Expect = 3e-32
Identities = 77/245 (31%), Positives = 113/245 (45%), Gaps = 2/245 (0%)
Frame = -2
Query: 41 VNTLGSWEHGRWHWKFLWRRPLVGRELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGIY 100
+ LG+ + W + WRRP E+ L + Q+ D W+W P G Y
Sbjct: 843 IQQLGTITNSGWE*QLNWRRPXXDSEIAMADSFLGEITQQQIHPQREDKWLWKPEPGGHY 664
Query: 101 SVNSAYHFL-QEPTLPDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITS 159
S S YH L E T D IW P+ FAWR++ DR ++ NL +RQV+
Sbjct: 663 STKSGYHVLWGELTEEIQDADFAEIWKLKIPTKAAVFAWRLVRDRLPTKSNLRRRQVM-- 490
Query: 160 LADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFRFGI-N 218
+ D C LCN+ E HL F+CT +L +W + W+ P + R H + I +
Sbjct: 489 VQDMVCPLCNNIEEGAAHLFFNCTKTLPLWWESMSWVNLKTAMPQTPRQHFLQYGTDIAD 310
Query: 219 KLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSS 278
L+ + W+A+ W++W RN+VVF+ + VLE W W KA K F+
Sbjct: 309 GLKSKRWKCWWIALTWTIWQHRNKVVFQNATFHGNKVLEDALLLLWSWFKALEKDFNLHF 130
Query: 279 SDWFS 283
+ W S
Sbjct: 129 NQWSS 115
>CO986021
Length = 900
Score = 127 bits (318), Expect = 8e-30
Identities = 74/241 (30%), Positives = 113/241 (46%), Gaps = 6/241 (2%)
Frame = -1
Query: 52 WHWKFLWRRPLVGRELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAYHFLQE 111
W+W F WRR L E E +D + + + + + D+ +W GIYS SAY L
Sbjct: 891 WNWDFKWRRNLFDHENEQAIAFMDDISAISIHQQLQDSMLWKADPTGIYSTKSAYRLLLP 712
Query: 112 PTLPDTDEALVSI-WHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCND 170
P + I W P + F+WR+ DR + R NLL+R V +L D C LC +
Sbjct: 711 NNRPGQHSSNFKILWKLKIPPRAELFSWRLFRDRLSIRANLLRRHV--ALQDIMCPLCGN 538
Query: 171 YVETGFHLLFSCTFSLSVWQGCLHW---LGCSFIQPSSARDHLSHFR--FGINKLQQRLA 225
+ E HL F C ++ +W ++W LG P++ H F FG + R
Sbjct: 537 HQEEAGHLFFHCRMTIGLWWESMNWTRTLGAFLDDPAA---HFIQFSDGFGAQRNHNRRC 367
Query: 226 LSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSNP 285
L W+A+ ++W RN ++F+G V++ +W W KA KGF+ + W +N
Sbjct: 366 L-WWIALTSTIWQHRNSLLFKGTPFQPPKVMDDALFHAWTWLKATEKGFNIPFNQWSTNL 190
Query: 286 L 286
L
Sbjct: 189 L 187
>TC213061
Length = 823
Score = 125 bits (313), Expect = 3e-29
Identities = 70/236 (29%), Positives = 106/236 (44%), Gaps = 3/236 (1%)
Frame = +2
Query: 52 WHWKFLWRRPLVGRELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAYHFLQE 111
W W+F WRRPL E+E + + ++ + D WVW + G Y SAY ++E
Sbjct: 20 WEWQFQWRRPLFDSEIEMLVAFIQEVEGHKIRSDIEDQWVWAAESSGSYLARSAYRVIRE 199
Query: 112 PTLP--DTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCN 169
+P + D +W P V FAWR+L D +R+NL +++V L + C LC
Sbjct: 200 -GIPEEEQDREFKELWKLKVPMKVTMFAWRLLKDILPTRDNLRRKRV--ELHEYVCPLCR 370
Query: 170 DYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHL-SHFRFGINKLQQRLALSI 228
E+ HL F C+ L +W L W+ P R H H LQ
Sbjct: 371 SMDESASHLFFHCSKILPIWWESLSWVKLVGAFPHHPRHHFHQHSHEVYQGLQGNRWKWW 550
Query: 229 WLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSN 284
WLA+ W++W RN+++F N + V++ W W + F+ + W SN
Sbjct: 551 WLALTWTIWKHRNDIIFSNATFNAHKVMDDAVFLIWTWFRHLENDFTSHYNQWSSN 718
>TC231922
Length = 803
Score = 116 bits (290), Expect = 1e-26
Identities = 63/225 (28%), Positives = 93/225 (41%), Gaps = 2/225 (0%)
Frame = -3
Query: 7 GDVTDFWHDDWLGTGC-LRGAFYRLYNLSRQKWNCVNTLGSWEHGRWHWKFLWRRPLVGR 65
GD FW D WL GC L+ + +LY +SRQ ++ +G + W+F WRR L
Sbjct: 690 GDKIKFWQDSWLSEGCNLQQKYNQLYTISRQ*NLTISKMGKFSQNARSWEFKWRRRLFDY 511
Query: 66 ELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAYHFLQEPTLPDTDEALVSI- 124
E ++ + + + D+ W + G+YS SAY L P + I
Sbjct: 510 EYAMAVDFMNEISGISIQNQAHDSMFWKADSSGVYSTKSAYRLLMPSISPAPSRRIFQIL 331
Query: 125 WHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTF 184
WH P F+WR+ LDR +R NL +R + + D C LC E HL F C
Sbjct: 330 WHLKIPPRAAVFSWRLFLDRLPTRGNLSRRSI--PIQDIMCPLCGCQHEEAGHLFFHCKM 157
Query: 185 SLSVWQGCLHWLGCSFIQPSSARDHLSHFRFGINKLQQRLALSIW 229
+ +W + W+ P+ H F G +W
Sbjct: 156 TKGLWWESMRWIRVIGALPADPASHFIQFCDGFAATSNHSRRGMW 22
>TC233392
Length = 1145
Score = 113 bits (283), Expect = 9e-26
Identities = 66/206 (32%), Positives = 94/206 (45%), Gaps = 2/206 (0%)
Frame = -1
Query: 81 QLTKGVPDAWVWMPSNVGIYSVNSAYHFLQEPTL-PDTDEALVSIWHSFAPSNVKGFAWR 139
Q+ + VPD WVW +YS SAY LQ D D AL +W P+ FAWR
Sbjct: 677 QIQQQVPDTWVWKHEPNELYSTRSAYKLLQGHLEGEDQDGALQDLWKLKIPAKASIFAWR 498
Query: 140 VLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCS 199
++ DR ++ NL +RQV+ L D+ C C E H+ C +W W+
Sbjct: 497 LIRDRLPTKSNLHRRQVV--LEDSLCPFCRIREEDASHIFLECNKIRPLWWESQTWVRSL 324
Query: 200 FIQPSSARDH-LSHFRFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLEL 258
+ P + R H L H + + W+A+ WS+W RN+V+F+ N +LE
Sbjct: 323 GVSPINKRQHFLQHVHGTPGSKRYNRWKTWWIALTWSLWKHRNQVIFQNAQFNGIKLLED 144
Query: 259 VKRRSWQWNKANMKGFSYSSSDWFSN 284
W W +A K FS + W SN
Sbjct: 143 AVFLHWTWIRAMEKDFSMHYNQWSSN 66
>TC231841
Length = 791
Score = 107 bits (267), Expect = 6e-24
Identities = 58/199 (29%), Positives = 88/199 (44%), Gaps = 2/199 (1%)
Frame = +3
Query: 88 DAWVWMPSNVGIYSVNSAYHFLQEPTLPDTDEALVS-IWHSFAPSNVKGFAWRVLLDRFA 146
D WVW G Y+ SAY L + + + +W PS + FAWR++ DR
Sbjct: 63 DQWVWKADPSGQYTAKSAYGVLWGEMFEEQQDGVFEELWKLKLPSKITIFAWRLIRDRLP 242
Query: 147 SRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSA 206
+R NL ++Q+ + D +C C E+ HL F C+ VW L W+ + P+
Sbjct: 243 TRSNLRRKQI--EVDDPRCPFCRSAEESAAHLFFHCSRIAPVWWESLSWVNLLGVFPNHP 416
Query: 207 RDHLSHFRFGINKLQQRLALSIW-LAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQ 265
R H +G+ + W LA+ W++W RN ++F G N +L+ W
Sbjct: 417 RQHFLQHIYGVTAGMRASRWKWWWLALTWTIWKQRNNMIFSNGTFNANKILDEAIFLIWT 596
Query: 266 WNKANMKGFSYSSSDWFSN 284
W K FS + W SN
Sbjct: 597 WLTHLEKDFSIHYNQWSSN 653
>TC233956
Length = 757
Score = 105 bits (262), Expect = 2e-23
Identities = 60/218 (27%), Positives = 97/218 (43%), Gaps = 3/218 (1%)
Frame = +2
Query: 70 EKGLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAYHFLQEPTLPDTDEAL--VSIWHS 127
E +D + ++L D WVW + GI S SAY ++ L D + L +W
Sbjct: 5 EAKFIDHIHEIRLNNNFNDTWVWRAESTGIISTKSAYQVIKSE-LDDEGQHLGFKKLWEI 181
Query: 128 FAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLS 187
P F WR+L DR +++NL+KRQ+ + D C C+ ET HL F+C +
Sbjct: 182 KVPPKALSFVWRLLWDRLPTKDNLIKRQI--QVEDDLCPFCHSQSETASHLFFTCGKIMP 355
Query: 188 VWQGCLHWLGCSFIQPSSARDH-LSHFRFGINKLQQRLALSIWLAVIWSVWLARNEVVFR 246
+W L W+ + D+ L H+ +K+ W+AV S+W RN+++F+
Sbjct: 356 LWWEFLSWVKEDKVFHCRPMDNFLQHYSSAASKVSNTRRTMWWIAVTNSIWRLRNDIIFQ 535
Query: 247 GGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSN 284
+I + + W W + + F W SN
Sbjct: 536 NQTVDITRLTDSTLFLLWTWLRGWERNFQIPYYCWSSN 649
>CO984521
Length = 716
Score = 100 bits (250), Expect = 6e-22
Identities = 61/221 (27%), Positives = 94/221 (41%), Gaps = 2/221 (0%)
Frame = -2
Query: 66 ELEWEKGLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAYHFLQEPTL-PDTDEALVSI 124
E++ L L + + D W W G YS SAY L T+ + D +
Sbjct: 712 EIDMAVAFLQQLEGFTIRPELSDQWKWAAEPSGCYSTKSAYKALHHVTVGEEQDGKFKEL 533
Query: 125 WHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTF 184
W P V FAWR++ D+ ++ NL K++V L + C LC ET HL F C+
Sbjct: 532 WKLRVPLKVAIFAWRLIQDKLPTKANLRKKRV--ELQEYLCPLCRSVEETASHLFFHCSK 359
Query: 185 SLSVWQGCLHWLGCSFIQPSSARDHLSHFRFGINK-LQQRLALSIWLAVIWSVWLARNEV 243
+W W+ + P H S FG + LQ + W A+ +S+W RN +
Sbjct: 358 VSPLWWESQSWVNMMGVFPYQPDQHFSQHIFGASVGLQGKRWQWWWFALTYSIWKHRNSI 179
Query: 244 VFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSN 284
+F + + +++ W W + K FS + W SN
Sbjct: 178 IFSNANFDAHKLMDDAVFILWTWLRCFEKDFSLHFNQWSSN 56
>TC213759
Length = 681
Score = 99.4 bits (246), Expect = 2e-21
Identities = 58/161 (36%), Positives = 84/161 (52%)
Frame = +2
Query: 129 APSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSV 188
AP V F W+V+L+R +RE L R + AD C +C+ T HL FSC F SV
Sbjct: 8 APLKVIVFGWQVILNRTQTREALF*RNIGGVDADLSCPMCHGDDVTTTHL-FSCRFGYSV 184
Query: 189 WQGCLHWLGCSFIQPSSARDHLSHFRFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGG 248
WQ C LG + P S+ + H + G ++ WL VIW +WL RN ++FRG
Sbjct: 185 WQRCYQRLGFEQVLPGSSCAYFWHHQ-GYGHW-----MTFWLVVIWLIWLQRNTIIFRGK 346
Query: 249 AANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSNPLLCI 289
A + V++ V ++W W KA ++S SD NP++ +
Sbjct: 347 QACVQEVVDSVVFKTWSWVKAYYARDTFSFSD*LLNPVISL 469
>BE660043
Length = 714
Score = 68.2 bits (165), Expect(2) = 4e-21
Identities = 36/113 (31%), Positives = 56/113 (48%), Gaps = 1/113 (0%)
Frame = -2
Query: 72 GLLDALGTVQLTKGVPDAWVWMPSNVGIYSVNSAYHFLQ-EPTLPDTDEALVSIWHSFAP 130
G L+ + L + D+WVW P + G YS +AY LQ + D+ +W P
Sbjct: 674 GFLEDISHTALQQHAADSWVWKPESNGYYSSRNAYILLQGNSEEGNMDDIFKDLWKLKIP 495
Query: 131 SNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCT 183
+ FA R++ DR ++ N+ KRQ+ + D+ C C+ ET HL F C+
Sbjct: 494 AKASIFAGRLIRDRLPTKSNMRKRQI--DINDSLCPFCSIKEETASHLFFDCS 342
Score = 50.4 bits (119), Expect(2) = 4e-21
Identities = 27/97 (27%), Positives = 45/97 (45%), Gaps = 1/97 (1%)
Frame = -1
Query: 189 WQGCLHWLGCSFIQPSSARDHLSHFRFGINKLQQRLALSIW-LAVIWSVWLARNEVVFRG 247
W L W+G S P + R H G+ ++ W +A+ WS+W RN+++F
Sbjct: 327 WWESLSWIGTSGAYPINPRLHFLQHNNGMKGGKKYNRWKCWWVALNWSIWQHRNKIIFTN 148
Query: 248 GAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSN 284
N ++LE +W+W + K F+ + W SN
Sbjct: 147 APFNGSNLLEDALFLAWKWFRTMEKNFTMHFNYWSSN 37
>CA851073
Length = 633
Score = 97.8 bits (242), Expect = 5e-21
Identities = 61/211 (28%), Positives = 96/211 (44%), Gaps = 6/211 (2%)
Frame = +1
Query: 80 VQLTKGVPDAWVWMPSNVGIYSVNSAYHFLQEPTLPDTDEALV-SIWHSFAPSNVKGFAW 138
+Q+ + D VW G+YS SAY L ++ + +IW P F+W
Sbjct: 4 IQIQSHLQDTMVWKADPCGVYSTKSAYRILMTCNRHVSEANIFKTIWKLKIPPRAAVFSW 183
Query: 139 RVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHW--- 195
R++ DR +R NLL+R V S+ + +C LC E HL F+C + +W + W
Sbjct: 184 RLIKDRLPTRHNLLRRNV--SIQENECPLCGYEQEEADHLFFNCKMTRGLWWESMRWNQI 357
Query: 196 LGCSFIQPSSARDHLSHF--RFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIY 253
+G + P+S H F FG + R W+A+ S+W RN ++F+G
Sbjct: 358 VGPLSVSPAS---HFVQFCDGFGAGRNHTRWC-GWWIALTSSIWKHRNLLIFQGNQFEPS 525
Query: 254 SVLELVKRRSWQWNKANMKGFSYSSSDWFSN 284
V++ +W W K K F+ + W SN
Sbjct: 526 KVMDDALFLAWSWLKVREKNFNTPFNHWSSN 618
>TC222789
Length = 954
Score = 92.8 bits (229), Expect = 2e-19
Identities = 49/168 (29%), Positives = 77/168 (45%), Gaps = 1/168 (0%)
Frame = +2
Query: 118 DEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFH 177
D+A +W P + FAWR+L DR ++ NL +RQ+ + D C C + E H
Sbjct: 344 DKAFEKLWKLKVPIKYEVFAWRLLRDRLPTKVNLHRRQI--QVMDRSCPFCRNVEEDAGH 517
Query: 178 LLFSCTFSLSVWQGCLHWLGCSFIQPSSARDH-LSHFRFGINKLQQRLALSIWLAVIWSV 236
L F C+ + +W L W+ S P R H L H ++ WLAV W++
Sbjct: 518 LFFHCSKIIPIWWESLSWVNISGALPKDPRQHFLQHDLIMAGGIRTTRWKCWWLAVTWTI 697
Query: 237 WLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSN 284
W RN+++F + + +++ W W K FS + + W SN
Sbjct: 698 WQQRNKIIFFNDSFDANKLIDEAAFLLWTWLSNLEKDFSVNFNQWSSN 841
>CD396188 weakly similar to GP|6175788|gb|A ORF144 {Xestia c-nigrum
granulovirus}, partial (5%)
Length = 701
Score = 89.7 bits (221), Expect = 1e-18
Identities = 53/193 (27%), Positives = 88/193 (45%), Gaps = 2/193 (1%)
Frame = -1
Query: 100 YSVNSAYHFLQEPTL-PDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVIT 158
+S NSAY+ ++ L + +W P FAWR+L DR S+ENL++RQ++
Sbjct: 701 FSTNSAYNCIRPDQLFYQPNSGFRQLWEIKIPPTALSFAWRLLWDRLPSKENLIRRQIV- 525
Query: 159 SLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDH-LSHFRFGI 217
L + C C VE+ HL F+C + +W W+ + S D+ L H
Sbjct: 524 -LQNDLCPFCQSQVESASHLFFTCHKVMPLWWEFNTWVREDRVLHSKPMDNFLQHCSLAG 348
Query: 218 NKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYS 277
++ R W+A S+W RN+++F +I +++ +W W + K F+
Sbjct: 347 SRNSNRRRKIWWIAATRSIWNLRNDMIFNNQPFDISKLVDKAIFLTWSWLRGWEKDFNVP 168
Query: 278 SSDWFSNPLLCIL 290
W SN L +
Sbjct: 167 FHQWSSNMSLSFI 129
>TC231904 similar to GB|AAL25568.1|16648753|AY058152 AT5g16150/T21H19_70
{Arabidopsis thaliana;} , partial (17%)
Length = 903
Score = 87.8 bits (216), Expect = 5e-18
Identities = 51/158 (32%), Positives = 79/158 (49%), Gaps = 4/158 (2%)
Frame = +1
Query: 131 SNVKG--FAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSV 188
SNV G AWR++ DR ++ NL +RQ+ ++D+ C C+ ET HL F C+ +
Sbjct: 256 SNVFGTIVAWRLIRDRLPTKSNLRRRQI--DISDSLCPFCSIKDETASHLFFECSKIQHL 429
Query: 189 WQGCLHWLGCSFIQPSSARDHLSHFRFGIN--KLQQRLALSIWLAVIWSVWLARNEVVFR 246
W W+G S P + R H G+ K+ +R W+A+ WS+W RN+V+F
Sbjct: 430 WWESQSWIGTSGAYPINPRHHFLQHNIGMKGGKIYKRWKCW-WVALNWSIWQQRNKVIFM 606
Query: 247 GGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSN 284
N +LE +W W +A + F+ + W SN
Sbjct: 607 NAPFNGSKLLEDALFVAWTWFRAMEENFTIHFNFWSSN 720
>TC222857
Length = 657
Score = 87.4 bits (215), Expect = 7e-18
Identities = 50/176 (28%), Positives = 75/176 (42%), Gaps = 1/176 (0%)
Frame = -3
Query: 110 QEPTLPDTDEALVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCN 169
+E + + + +W PS FAWR+L DR ++ NL RQV ++D C C
Sbjct: 655 EEVAVDNLHDCFKDLWKIKIPSKFLMFAWRLLWDRLPTKANLRARQV--QISDLTCPFCR 482
Query: 170 DYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFR-FGINKLQQRLALSI 228
ET H+ C + +W ++W+ P S DH F ++ R
Sbjct: 481 RVEETASHMFIHCIKTQPIWWETMNWINMQGPLPWSITDHFMQFSSLKEAGIRSRRWQWW 302
Query: 229 WLAVIWSVWLARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSN 284
W+AV WS+W RN +VF + ++E W W K FS + W SN
Sbjct: 301 WMAVTWSIWQLRNSIVFSNATFDGNKLVEDASFLLWTWLHNLEKDFSLHFNQWSSN 134
>TC210600 similar to UP|Q51534 (Q51534) PilW protein, partial (7%)
Length = 1093
Score = 83.2 bits (204), Expect = 1e-16
Identities = 50/159 (31%), Positives = 73/159 (45%), Gaps = 10/159 (6%)
Frame = +3
Query: 143 DRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLFSCTFSLSVWQGCLHWLGCSFIQ 202
DR +R NL RQV L D++C LCN E HL F CT +L +W W+
Sbjct: 519 DRLPTRTNLRVRQV--ELNDSRCPLCNSLEEDAAHLFFPCTKTLPLWWETQSWINSLGAF 692
Query: 203 PSSARDH-LSHFRFGINKLQQRLALSIWLAVIWSVWLARNEVVFRGGAANIYSVLELVKR 261
P + +DH L H + ++ R W+A+ W +W RN VVF + + V++
Sbjct: 693 PHNPKDHFLQHDNSSADGVRARRWKCWWVALTWVIWKHRNGVVFHNHSFDGSKVMDDAIF 872
Query: 262 RSWQWNKANMKGFSYSSSDW---------FSNPLLCILG 291
W W K KGF+ S W F ++C++G
Sbjct: 873 LFWSWLKVMEKGFTVPFSSWSSQLKAAF*F*GVIICVIG 989
>TC233056
Length = 680
Score = 82.4 bits (202), Expect = 2e-16
Identities = 48/167 (28%), Positives = 83/167 (48%), Gaps = 3/167 (1%)
Frame = -1
Query: 121 LVSIWHSFAPSNVKGFAWRVLLDRFASRENLLKRQVITSLADAQCALCNDYVETGFHLLF 180
+V +W P+ FAWR++ DR ++ NL +RQV+ L D+ C C + E HL F
Sbjct: 668 IVELWKLKIPAKSAVFAWRLIRDRLPTKTNLRRRQVM--LTDSLCPFCRNKEEEATHLFF 495
Query: 181 SCTFSLSVWQGCLHWLGCSFIQPSSARDHLSHFRFGINKLQQRLALSI---WLAVIWSVW 237
+ + L +W L + + P + RDH + + GI + A+ W+A+ ++W
Sbjct: 494 NYSNILPLWWESLS*VNLTTALPQNPRDH--YLQHGIGNDHGKKAIRWKCWWIALTRTIW 321
Query: 238 LARNEVVFRGGAANIYSVLELVKRRSWQWNKANMKGFSYSSSDWFSN 284
RN++VF+ + + +L+ W W K+ K F+ + W SN
Sbjct: 320 QHRNKIVFQNASFDGSKLLDDAVLLIWTWVKSMEKDFAVHFNQWSSN 180
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.138 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,687,480
Number of Sequences: 63676
Number of extensions: 367132
Number of successful extensions: 3286
Number of sequences better than 10.0: 147
Number of HSP's better than 10.0 without gapping: 3125
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3189
length of query: 291
length of database: 12,639,632
effective HSP length: 96
effective length of query: 195
effective length of database: 6,526,736
effective search space: 1272713520
effective search space used: 1272713520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0303.2


