

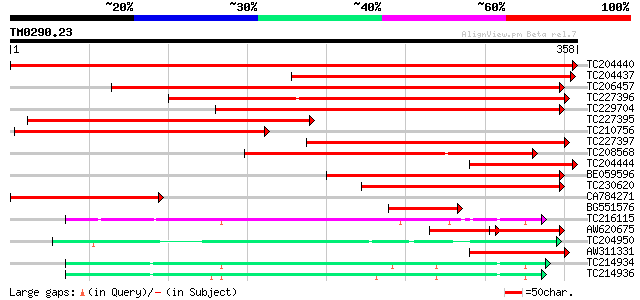
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0290.23
(358 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204440 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehy... 664 0.0
TC204437 homologue to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol de... 330 5e-91
TC206457 weakly similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannit... 269 1e-72
TC227396 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehy... 248 2e-66
TC229704 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehy... 220 7e-58
TC227395 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehy... 208 3e-54
TC210756 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehy... 181 5e-46
TC227397 similar to UP|MTD1_STYHU (Q43137) Probable mannitol deh... 157 5e-39
TC208568 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {A... 143 1e-34
TC204444 homologue to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol de... 131 4e-31
BE059596 120 1e-27
TC230620 similar to UP|Q9ATW1 (Q9ATW1) Cinnamyl alcohol dehydrog... 118 5e-27
CA784271 104 7e-23
BG551576 homologue to SP|P30359|CAD4_ Cinnamyl-alcohol dehydroge... 87 9e-18
TC216115 weakly similar to UP|ADHX_UROHA (P80467) Alcohol dehydr... 75 6e-14
AW620675 similar to GP|13507210|gb| cinnamyl alcohol dehydrogena... 52 7e-14
TC204950 similar to UP|Q945P3 (Q945P3) At1g23740/F5O8_27, partia... 69 4e-12
AW311331 66 2e-11
TC214934 homologue to UP|Q43016 (Q43016) Alcohol dehydrogenase-1... 62 5e-10
TC214936 UP|O82478 (O82478) Alcohol dehydrogenase Adh-1 (Fragme... 60 2e-09
>TC204440 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (97%)
Length = 1481
Score = 664 bits (1712), Expect = 0.0
Identities = 320/358 (89%), Positives = 342/358 (95%)
Frame = +3
Query: 1 MGSLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGM 60
MGSLEAERTTVG AARDPSGILSPYT+ LRNTGPDDVYIKVHYCG+CH+D+HQ+KNDLGM
Sbjct: 60 MGSLEAERTTVGLAARDPSGILSPYTYNLRNTGPDDVYIKVHYCGICHSDLHQIKNDLGM 239
Query: 61 SNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWN 120
SNYPMVPGHEVVGEVLEVGSDV+RF VGE+VG GLLVGCCK C CQ DIE YC KKIW+
Sbjct: 240 SNYPMVPGHEVVGEVLEVGSDVSRFRVGELVGVGLLVGCCKNCQPCQQDIENYCSKKIWS 419
Query: 121 YNDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKES 180
YNDVYVDGKPTQGGFAET+IVEQKFVVKIPEG+APEQVAPLLCA VTVYSPL HFGLKES
Sbjct: 420 YNDVYVDGKPTQGGFAETMIVEQKFVVKIPEGLAPEQVAPLLCAGVTVYSPLVHFGLKES 599
Query: 181 GLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQE 240
GLRGGILGLGGVGHMGV IAKA+GHHVTVISSSD+KK+EA+E LGAD YLVSSD T+MQE
Sbjct: 600 GLRGGILGLGGVGHMGVKIAKALGHHVTVISSSDKKKQEALEHLGADQYLVSSDATAMQE 779
Query: 241 AADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFI 300
AADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQF++PMVMLGR+SITGSFI
Sbjct: 780 AADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFVSPMVMLGRKSITGSFI 959
Query: 301 GSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGSKLIDQ 358
GS+KETEEMLEFWKEKGL+SMIE+VNMDYINKAFERLEKNDVRYRFVVDVKGSKL+DQ
Sbjct: 960 GSMKETEEMLEFWKEKGLSSMIEMVNMDYINKAFERLEKNDVRYRFVVDVKGSKLVDQ 1133
>TC204437 homologue to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (49%)
Length = 844
Score = 330 bits (847), Expect = 5e-91
Identities = 164/179 (91%), Positives = 175/179 (97%)
Frame = +3
Query: 179 ESGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSM 238
ESGLRGGILGLGGVGHMGV IAKA+GHHVTVISSSD+KK+EA+E LGAD YLVSSD T+M
Sbjct: 3 ESGLRGGILGLGGVGHMGVKIAKALGHHVTVISSSDKKKQEALEHLGADQYLVSSDVTAM 182
Query: 239 QEAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGS 298
QEAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQF++PMVMLGRRSITGS
Sbjct: 183 QEAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFVSPMVMLGRRSITGS 362
Query: 299 FIGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGSKLID 357
FIGS+KETEEMLEFWKEKGL+SMIE+VNMDYINKAFERLEKNDVRYRFVVDVKGSKL+D
Sbjct: 363 FIGSMKETEEMLEFWKEKGLSSMIEVVNMDYINKAFERLEKNDVRYRFVVDVKGSKLVD 539
>TC206457 weakly similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol
dehydrogenase (NAD-dependent mannitol dehydrogenase) ,
partial (75%)
Length = 1180
Score = 269 bits (688), Expect = 1e-72
Identities = 138/288 (47%), Positives = 192/288 (65%), Gaps = 2/288 (0%)
Frame = +3
Query: 65 MVP-GHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYND 123
+VP GHE++G V +VG DV F G+ VG G L C +C C++D E YC+K + YN
Sbjct: 15 LVPNGHEIIGVVTKVGRDVKGFKEGDRVGVGCLAASCLECEHCKTDQENYCEKLQFVYNG 194
Query: 124 VYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKES-GL 182
V+ DG T GG+++ + + ++VV IPE +A + APLLCA +TV++PL L S G
Sbjct: 195 VFWDGSITYGGYSQIFVADYRYVVHIPENLAMDAAAPLLCAGITVFNPLKDHDLVASPGK 374
Query: 183 RGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAA 242
+ G++GLGG+GH+ V KA GHHVTVIS+S K+ EA + LGAD ++VSS+ +Q A
Sbjct: 375 KIGVVGLGGLGHIAVKFGKAFGHHVTVISTSPSKEAEAKQRLGADDFIVSSNPKQLQAAR 554
Query: 243 DSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGS 302
S+D+I+DTV H L P L LLK++G L L+G + PLQ ++ G+RS+ G IG
Sbjct: 555 RSIDFILDTVSAEHSLLPILELLKVNGTLFLVGAPDKPLQLPAFPLIFGKRSVKGGIIGG 734
Query: 303 IKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
IKET+EMLE + +TS IE++ D IN+A ERL KNDVRYRFV+D+
Sbjct: 735 IKETQEMLEVCAKYNITSDIELITPDRINEAMERLAKNDVRYRFVIDI 878
>TC227396 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (70%)
Length = 927
Score = 248 bits (634), Expect = 2e-66
Identities = 123/253 (48%), Positives = 173/253 (67%)
Frame = +2
Query: 101 KKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAP 160
K+C CQ D+E YC + ++ YN Y DG TQGG++ ++V Q++V++ PE + + AP
Sbjct: 2 KECENCQQDLENYCPRPVFTYNSPYYDGTRTQGGYSNIVVVHQRYVLRFPENLPLDAGAP 181
Query: 161 LLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEA 220
LLCA +TVYSP+ ++G+ E G+ GL G+GH+ + +AKA G VTVISSS K+ EA
Sbjct: 182 LLCAGITVYSPMKYYGMTEPA-NIGVEGLVGLGHVAIKLAKAFGLKVTVISSSPNKQAEA 358
Query: 221 IEDLGADAYLVSSDTTSMQEAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTP 280
I+ LGAD++LVSSD M+ A ++DYIIDT+ H L P L LLKL+GKL+ +G+ N P
Sbjct: 359 IDRLGADSFLVSSDPAKMKVALGTMDYIIDTISAVHSLIPLLGLLKLNGKLVTVGLPNKP 538
Query: 281 LQFITPMVMLGRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKN 340
L+ ++ GR+ I GS G IKET+EML+F + +T+ IE++ MD IN A ERL K
Sbjct: 539 LELPIFPLVAGRKLIGGSNFGGIKETQEMLDFCAKHNITADIELIKMDQINTAMERLSKA 718
Query: 341 DVRYRFVVDVKGS 353
DV+YRFV+DV S
Sbjct: 719 DVKYRFVIDVANS 757
>TC229704 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (63%)
Length = 816
Score = 220 bits (561), Expect = 7e-58
Identities = 106/220 (48%), Positives = 155/220 (70%)
Frame = +2
Query: 131 TQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLG 190
T GG++++++ ++ FVV+IP+ + + APLLCA +TVYSPL ++GL + GL G++GLG
Sbjct: 2 TYGGYSDSMVADEHFVVRIPDRLPLDAAAPLLCAGITVYSPLRYYGLDKPGLHVGVVGLG 181
Query: 191 GVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIID 250
G+GHM V AKA G VTVIS+S KK+EAI++LGAD++L+S D MQ A +LD IID
Sbjct: 182 GLGHMAVKFAKAFGAKVTVISTSPNKKEEAIQNLGADSFLISRDQDQMQAAMGTLDGIID 361
Query: 251 TVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEML 310
TV HPL P + LLK GKL+++G PL+ ++ GR+ + G+ IG + ET+EM+
Sbjct: 362 TVSAVHPLLPLIGLLKSHGKLVMVGAPEKPLELPVFPLLAGRKIVAGTLIGGLMETQEMI 541
Query: 311 EFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
+F + + IE++ MDY+N A ERL K DV+YRFV+D+
Sbjct: 542 DFAAKHNVKPDIEVIPMDYVNTAMERLLKADVKYRFVIDI 661
>TC227395 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (52%)
Length = 1002
Score = 208 bits (529), Expect = 3e-54
Identities = 89/181 (49%), Positives = 127/181 (69%)
Frame = +1
Query: 12 GWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEV 71
GWAA D SG L+P+ F+ R G DDV +K+ +CGVCH+D+H +KND G + YP+VPGHE+
Sbjct: 76 GWAASDTSGTLAPFHFSRRENGVDDVTLKILFCGVCHSDLHTLKNDWGFTTYPVVPGHEI 255
Query: 72 VGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPT 131
VG V EVG++V F VG+ VG G++V CK+C CQ D+E YC + ++ YN Y DG T
Sbjct: 256 VGVVTEVGNNVKNFKVGDKVGVGVIVESCKECENCQQDLENYCPRPVFTYNSPYYDGTRT 435
Query: 132 QGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGG 191
QGG++ ++V Q++V++ PE + + APLLCA +T YSP+ ++G+ E G G+ LGG
Sbjct: 436 QGGYSNIMVVHQRYVLRFPENLPLDAGAPLLCAGITXYSPMKYYGMTEPGKHLGVARLGG 615
Query: 192 V 192
V
Sbjct: 616 V 618
>TC210756 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (45%)
Length = 549
Score = 181 bits (459), Expect = 5e-46
Identities = 76/161 (47%), Positives = 109/161 (67%)
Frame = +3
Query: 4 LEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGMSNY 63
+E + GWAARD SG+LSP+ F+ R TG D+ KV YCG+CH+D+H +KN+ G + Y
Sbjct: 66 MEHPKKVFGWAARDSSGLLSPFNFSRRETGEKDLVFKVQYCGICHSDLHMLKNEWGNTTY 245
Query: 64 PMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYND 123
P+VPGHE+ G V EVGS V +F VG+ VG G ++G C+ C +C ++E YC K I Y
Sbjct: 246 PLVPGHEIAGVVTEVGSKVQKFKVGDRVGVGCMIGSCRSCESCDENLENYCPKMILTYGV 425
Query: 124 VYVDGKPTQGGFAETIIVEQKFVVKIPEGMAPEQVAPLLCA 164
Y DG T GG+++ ++ ++ FVV+IP+ + + APLLCA
Sbjct: 426 KYFDGTITHGGYSDLMVADEHFVVRIPDNLPLDAAAPLLCA 548
>TC227397 similar to UP|MTD1_STYHU (Q43137) Probable mannitol dehydrogenase 1
(NAD-dependent mannitol dehydrogenase 1) , partial
(48%)
Length = 742
Score = 157 bits (398), Expect = 5e-39
Identities = 84/166 (50%), Positives = 112/166 (66%)
Frame = +2
Query: 188 GLGGVGHMGVIIAKAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDY 247
GL G+ H +AKA G VTVISSS K+ EAI+ LGAD +LVSSD M+ A ++DY
Sbjct: 2 GLXGLXHXXXKLAKAFGLKVTVISSSPXKQAEAIDRLGADFFLVSSDPAKMKAALGTMDY 181
Query: 248 IIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETE 307
IIDT+ H L P L LLKL+GKL+ +G+ N PL+ ++ GR+ I GS G +KET+
Sbjct: 182 IIDTISAVHSLIPLLGLLKLNGKLVTVGLPNKPLELPIFPLVAGRKLIGGSNFGGLKETQ 361
Query: 308 EMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDVKGS 353
EML+F + +T+ IE++ MD IN A ERL + DV+YRFV+DV S
Sbjct: 362 EMLDFCGKHNITADIELIKMDQINTAMERLSRADVKYRFVIDVASS 499
>TC208568 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Arabidopsis
thaliana;} , partial (52%)
Length = 556
Score = 143 bits (361), Expect = 1e-34
Identities = 76/185 (41%), Positives = 112/185 (60%)
Frame = +1
Query: 149 IPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGHMGVIIAKAMGHHVT 208
IP+ APLLCA +TVYSP+ + + G G++GLGG+GHM V KA G VT
Sbjct: 4 IPKSYPLAAAAPLLCAGITVYSPMVRHKMNQPGKSLGVIGLGGLGHMAVKFGKAFGLSVT 183
Query: 209 VISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIIDTVPVGHPLEPYLSLLKLD 268
V S+S KK+EA+ LGAD ++VSS+ M A SLD+IIDT H +PY+SLLK
Sbjct: 184 VFSTSISKKEEALSLLGADKFVVSSNQEEMTALAKSLDFIIDTASGDHSFDPYMSLLKTY 363
Query: 269 GKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMD 328
G +L+G + ++FI + +G +++ GS G K+ +EM+ F + IE++ ++
Sbjct: 364 GVFVLVG-FPSQVKFIPASLNIGSKTVAGSVTGGTKDIQEMIGFCAANEIHPNIEVIPIE 540
Query: 329 YINKA 333
Y N+A
Sbjct: 541 YANEA 555
>TC204444 homologue to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (22%)
Length = 790
Score = 131 bits (330), Expect = 4e-31
Identities = 63/68 (92%), Positives = 68/68 (99%)
Frame = +1
Query: 291 GRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
GR+SITGSFIGS+KETEEMLEFWKEKGL+SMIE+VNMDYINKAFERLEKNDVRYRFVVDV
Sbjct: 475 GRKSITGSFIGSMKETEEMLEFWKEKGLSSMIEMVNMDYINKAFERLEKNDVRYRFVVDV 654
Query: 351 KGSKLIDQ 358
KGSKL+DQ
Sbjct: 655 KGSKLVDQ 678
Score = 30.8 bits (68), Expect = 0.99
Identities = 12/14 (85%), Positives = 14/14 (99%)
Frame = +2
Query: 278 NTPLQFITPMVMLG 291
NTPLQF++PMVMLG
Sbjct: 2 NTPLQFVSPMVMLG 43
>BE059596
Length = 466
Score = 120 bits (300), Expect = 1e-27
Identities = 65/150 (43%), Positives = 95/150 (63%)
Frame = +2
Query: 201 KAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIIDTVPVGHPLEP 260
KA GHHVTVIS+ K+ EA + LGAD +++S + +Q A S+D+I+DTV H L P
Sbjct: 8 KAFGHHVTVISTCPSKEPEAKQRLGADHFILSCNPKQLQAARTSMDFILDTVVAEHSLLP 187
Query: 261 YLSLLKLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEMLEFWKEKGLTS 320
L LLK++G L+G + PLQ ++ G+RS+ G IG IK+T++MLE + + S
Sbjct: 188 ILELLKVNGT*FLVGAPDKPLQSPAFPLIFGKRSVKGGIIGGIKKTQDMLEVRAKYNIRS 367
Query: 321 MIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
IE+V I++A ERL ND R RF++ +
Sbjct: 368 DIELVTPHTIDEAMERLAHNDERCRFMLGI 457
>TC230620 similar to UP|Q9ATW1 (Q9ATW1) Cinnamyl alcohol dehydrogenase,
partial (37%)
Length = 690
Score = 118 bits (295), Expect = 5e-27
Identities = 55/128 (42%), Positives = 87/128 (67%)
Frame = +1
Query: 223 DLGADAYLVSSDTTSMQEAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQ 282
++GAD+++VS + MQ ++D IIDTV HPL P + LLK GKL+++G PL+
Sbjct: 7 NIGADSFVVSREQDQMQAVMGTMDGIIDTVSAVHPLVPLIGLLKPHGKLVMVGAPEKPLE 186
Query: 283 FITPMVMLGRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDV 342
+++GR+ + GS IG +KET+EM++F + G+ IE++ +DY+N A ERL K D+
Sbjct: 187 LPVFSLLMGRKMVGGSSIGGMKETQEMIDFAAKHGVKPDIEVIPIDYVNTAIERLAKADL 366
Query: 343 RYRFVVDV 350
+YRFV+D+
Sbjct: 367 KYRFVIDI 390
>CA784271
Length = 436
Score = 104 bits (259), Expect = 7e-23
Identities = 51/97 (52%), Positives = 63/97 (64%)
Frame = +3
Query: 1 MGSLEAERTTVGWAARDPSGILSPYTFTLRNTGPDDVYIKVHYCGVCHTDVHQVKNDLGM 60
M S +GWAARD SG+LSPY F+ R G +DV IK+ +CGVC DV +N G
Sbjct: 144 MSSKGVGEDCLGWAARDASGVLSPYKFSRRTPGNEDVLIKITHCGVCFADVVWTRNKHGD 323
Query: 61 SNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGLLV 97
S YP+VPGHE+ G V +VGS+V RF VG+ VG G V
Sbjct: 324 SKYPVVPGHEIAGIVTKVGSNVHRFKVGDHVGVGTYV 434
>BG551576 homologue to SP|P30359|CAD4_ Cinnamyl-alcohol dehydrogenase (EC
1.1.1.195) (CAD). [Common tobacco] {Nicotiana tabacum},
partial (14%)
Length = 352
Score = 87.4 bits (215), Expect = 9e-18
Identities = 42/47 (89%), Positives = 43/47 (91%)
Frame = +1
Query: 240 EAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFITP 286
EAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTP + P
Sbjct: 19 EAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPFILLAP 159
>TC216115 weakly similar to UP|ADHX_UROHA (P80467) Alcohol dehydrogenase
class III (Glutathione-dependent formaldehyde
dehydrogenase) (FDH) , partial (42%)
Length = 1433
Score = 74.7 bits (182), Expect = 6e-14
Identities = 81/335 (24%), Positives = 135/335 (40%), Gaps = 31/335 (9%)
Frame = +1
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGL 95
+V +K+ +C TD+ K +N+P+ GHE VG + VG VT G++V
Sbjct: 166 EVRVKMLCASICSTDISSTKG-FPHTNFPIALGHEGVGIIESVGDQVTNLKEGDVV-IPT 339
Query: 96 LVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQ-------------GGFAETIIVE 142
+G C++C C S+ C + + D ++E ++ +
Sbjct: 340 YIGECQECENCVSEKTNLCMTYPVRWTGLMPDNTSRMSIRGERIYHIFSCATWSEYMVSD 519
Query: 143 QKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGHMGVIIAKA 202
+V+K+ + + + C T + ESG + GLG VG VI +K
Sbjct: 520 ANYVLKVDPTIDRAHASFISCGFSTGFGAAWKEAKVESGSTVAVFGLGAVGLGAVIGSKM 699
Query: 203 MGHHVTVISSSDRKKKEAIEDLGADAYLVSSDTT-SMQEAADSL------DYIIDTVPVG 255
G + ++ K+ E G ++ D+ S E L DY + V
Sbjct: 700 QGASRIIGIDTNENKRAKGEAFGITDFINPGDSNKSASELVKELSGGMGADYSFECTGVS 879
Query: 256 HPLEPYLSLLKL-DGKLILMGV---INTPLQFITPMVMLGRRSITGSFIGSIKETEEMLE 311
L L K+ GK I++GV I PL ++LG R++ GS G ++ + L
Sbjct: 880 TLLSESLEATKIGTGKAIVIGVGIEITLPLGLFA--ILLG-RTLKGSVFGGLRAISD-LS 1047
Query: 312 FWKEKGLTSMIEI-------VNMDYINKAFERLEK 339
+KG + V + INKAFE L++
Sbjct: 1048ILADKGHKKEFPLQELFTHEVTLADINKAFELLKQ 1152
>AW620675 similar to GP|13507210|gb| cinnamyl alcohol dehydrogenase {Fragaria
x ananassa}, partial (24%)
Length = 406
Score = 52.4 bits (124), Expect(2) = 7e-14
Identities = 22/47 (46%), Positives = 34/47 (71%)
Frame = +2
Query: 304 KETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
+ET+E+++F E + IE++ MDY+N A ERL DV+YRFV+D+
Sbjct: 122 QETQEIIDFATEHNVRPQIEVIPMDYVNIAMERLLNADVKYRFVIDI 262
Score = 42.4 bits (98), Expect(2) = 7e-14
Identities = 16/45 (35%), Positives = 31/45 (68%)
Frame = +3
Query: 266 KLDGKLILMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEML 310
K GKL+++G PL+ + P +++GR++I S+IG +K +++L
Sbjct: 9 KSHGKLVMVGAPEKPLELLLPSLIMGRKTIAASYIGGVKRHKKLL 143
>TC204950 similar to UP|Q945P3 (Q945P3) At1g23740/F5O8_27, partial (80%)
Length = 1560
Score = 68.6 bits (166), Expect = 4e-12
Identities = 77/324 (23%), Positives = 127/324 (38%), Gaps = 3/324 (0%)
Frame = +1
Query: 28 TLRNTGPDDVYIKVHYCGVCHTDV--HQVKNDLGMSNYPMVPGHEVVGEVLEVGSDVTRF 85
T+ + D V IKV + D Q K S P VPG++V G V++VGS V F
Sbjct: 346 TVPDVKEDQVLIKVVAAALNPVDAKRRQGKFKATDSPLPTVPGYDVAGVVVKVGSQVKDF 525
Query: 86 TVGEIVGAGLLVGCCKKCTACQSDIEQYCKKKIWNYNDVYVDGKPTQGGFAETIIVEQKF 145
VG+ V + N+ ++G G AE VE+K
Sbjct: 526 KVGDEVYGDV--------------------------NEKALEGPKQFGSLAEYTAVEEKL 627
Query: 146 VVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGHMGVIIAKAMGH 205
+ P+ + Q A L A T Y L G + G GGVG + + +AK +
Sbjct: 628 LAPKPKNLDFAQAASLPLAIETAYEGLERTGFSPGKSILVLNGSGGVGSLVIQLAKQVYG 807
Query: 206 HVTVISSSDRKKKEAIEDLGADAYLVSSDTTSMQEAADSLDYIIDTVPVGHPLEPYLSLL 265
V ++S + + ++ LGAD + + ++ + D + D + + + +
Sbjct: 808 ASRVAATSSTRNLDLLKSLGAD-LAIDYTKENFEDLPEKFDVVYDAI---GQCDRAVKAV 975
Query: 266 KLDGKLI-LMGVINTPLQFITPMVMLGRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEI 324
K DG ++ L G + P G R + S +++ LE K K +
Sbjct: 976 KEDGSVVALTGAVTPP----------GFRFVVTSNGDVLRKLNPYLESGKVKPIVDPKGP 1125
Query: 325 VNMDYINKAFERLEKNDVRYRFVV 348
+ D + +AF LE N + V+
Sbjct: 1126FSFDKLAEAFSYLETNRATGKVVI 1197
>AW311331
Length = 317
Score = 66.2 bits (160), Expect = 2e-11
Identities = 28/63 (44%), Positives = 46/63 (72%)
Frame = +1
Query: 291 GRRSITGSFIGSIKETEEMLEFWKEKGLTSMIEIVNMDYINKAFERLEKNDVRYRFVVDV 350
GR+ + GS IG +KET+EM++F + G+ IE++ +DY+N A +RL K DV+Y FV+D+
Sbjct: 4 GRKMVGGSSIGGMKETQEMIDFAAKHGVKPDIEVIPIDYVNTAIDRLAKADVKYLFVIDI 183
Query: 351 KGS 353
+ +
Sbjct: 184 ENT 192
>TC214934 homologue to UP|Q43016 (Q43016) Alcohol dehydrogenase-1F ,
complete
Length = 1450
Score = 61.6 bits (148), Expect = 5e-10
Identities = 79/336 (23%), Positives = 131/336 (38%), Gaps = 30/336 (8%)
Frame = +3
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGL 95
+V +K+ Y +CHTDV+ + +P + GHE G V VG VT G+ +
Sbjct: 201 EVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTHLKPGD-HALPV 377
Query: 96 LVGCCKKCTACQSDIEQYCK-KKIWNYNDVYVDGKPTQ--------------GGFAETII 140
G C +C C+S+ C +I V + T+ F+E +
Sbjct: 378 FTGECGECPHCKSEESNMCDLLRINTDRGVMIHDSQTRFSIKGQPIYHFVGTSTFSEYTV 557
Query: 141 VEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGHMGVIIA 200
V V K+ +++ L C T + + G I GLG VG A
Sbjct: 558 VHAGCVAKVNPAAPLDKICVLSCGICTGLGATINVAKPKPGSSVAIFGLGAVGLAAAEGA 737
Query: 201 KAMGHHVTVISSSDRKKKEAIEDLGADAYLVSSD-TTSMQE--AADSLDYIIDTVPVGHP 257
+ G + + E + G + ++ D +QE AA + + V
Sbjct: 738 RISGASRIIGVDLVSSRFEEAKKFGVNEFVNPKDHDKPVQEVIAAMTNGGVDRAVECTGS 917
Query: 258 LEPYLSLLKLD----GKLILMGVINTPLQFIT-PMVMLGRRSITGSFIGSIKETEEMLEF 312
++ +S + G +L+GV N F T P+ L R++ G+F G+ K + L
Sbjct: 918 IQAMISAFECVHDGWGVAVLVGVPNKDDAFKTHPVNFLNERTLKGTFYGNYKPRTD-LPS 1094
Query: 313 WKEKGLTSMIEI-------VNMDYINKAFERLEKND 341
EK + +E+ V INKAF+ + K +
Sbjct: 1095VVEKYMNGELELEKFITHTVPFSEINKAFDYMLKGE 1202
>TC214936 UP|O82478 (O82478) Alcohol dehydrogenase Adh-1 (Fragment) ,
complete
Length = 1353
Score = 59.7 bits (143), Expect = 2e-09
Identities = 79/334 (23%), Positives = 131/334 (38%), Gaps = 30/334 (8%)
Frame = +3
Query: 36 DVYIKVHYCGVCHTDVHQVKNDLGMSNYPMVPGHEVVGEVLEVGSDVTRFTVGEIVGAGL 95
+V +K+ Y +CHTDV+ +P + GHE G V VG VT G+ +
Sbjct: 138 EVRLKILYTSLCHTDVYFWDAKGQTPLFPRIFGHEASGIVESVGEGVTHLKPGD-HALPV 314
Query: 96 LVGCCKKCTACQSDIEQYCK-KKIWNYNDVYV---------DGKPTQ-----GGFAETII 140
G C C C+S+ C+ +I V + +G+P F+E +
Sbjct: 315 FTGECGDCAHCKSEESNMCELLRINTDRGVMIHDGQSRFSKNGQPIHHFLGTSTFSEYTV 494
Query: 141 VEQKFVVKIPEGMAPEQVAPLLCAAVTVYSPLSHFGLKESGLRGGILGLGGVGHMGVIIA 200
V V KI ++V L C T + + + G I GLG V A
Sbjct: 495 VHAGCVAKINPAAPLDKVCVLSCGICTGFGATVNVAKPKPGSSVAIFGLGAVALAAAEGA 674
Query: 201 KAMGHHVTVISSSDRKKKEAIEDLGADAYL--VSSDTTSMQEAADSLDYIID-TVPVGHP 257
+ G + + E + G + ++ D Q A+ + +D V
Sbjct: 675 RVSGASRIIGVDLVSARFEEAKKFGVNEFVNPKDHDKPVQQVIAEMTNGGVDRAVECTGS 854
Query: 258 LEPYLSLLKLD----GKLILMGVINTPLQFIT-PMVMLGRRSITGSFIGSIKETEEMLEF 312
++ +S + G +L+GV + F T P+ L R++ G+F G+ K + L
Sbjct: 855 IQAMVSAFECVHDGWGLAVLVGVPSKDDAFKTAPINFLNERTLKGTFYGNYKPRTD-LPS 1031
Query: 313 WKEKGLTSMIEI-------VNMDYINKAFERLEK 339
EK ++ +E+ V INKAF+ + K
Sbjct: 1032VVEKYMSGELEVDKFITHTVPFSEINKAFDLMLK 1133
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.139 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,945,754
Number of Sequences: 63676
Number of extensions: 196669
Number of successful extensions: 987
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 974
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 981
length of query: 358
length of database: 12,639,632
effective HSP length: 98
effective length of query: 260
effective length of database: 6,399,384
effective search space: 1663839840
effective search space used: 1663839840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0290.23


