

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0290.12
(143 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
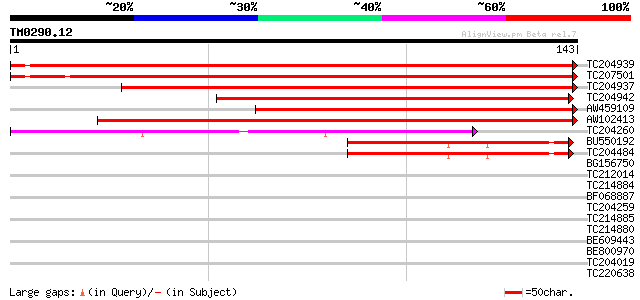
Score E
Sequences producing significant alignments: (bits) Value
TC204939 similar to GB|AAN38679.1|23507751|BT000842 At4g34190/F2... 210 2e-55
TC207501 similar to GB|AAN38679.1|23507751|BT000842 At4g34190/F2... 185 7e-48
TC204937 similar to GB|AAN38679.1|23507751|BT000842 At4g34190/F2... 183 2e-47
TC204942 homologue to UP|Q6W5F3 (Q6W5F3) Microtubule-associated ... 142 5e-35
AW459109 128 1e-30
AW102413 117 2e-27
TC204260 similar to UP|PSBS_SPIOL (Q02060) Photosystem II 22 kDa... 43 6e-05
BU550192 homologue to PIR|JC5876|JC5 early light-inducible prote... 40 5e-04
TC204484 homologue to UP|P93169 (P93169) Early light-induced pro... 40 5e-04
BG156750 similar to SP|P54773|PSBS Photosystem II 22 kDa protein... 38 0.002
TC212014 similar to UP|PSBS_ARATH (Q9XF91) Photosystem II 22 kDa... 37 0.003
TC214884 similar to UP|P93169 (P93169) Early light-induced prote... 37 0.003
BF068887 homologue to PIR|JC5876|JC58 early light-inducible prot... 37 0.003
TC204259 similar to UP|PSBS_SPIOL (Q02060) Photosystem II 22 kDa... 36 0.005
TC214885 homologue to UP|P93169 (P93169) Early light-induced pro... 36 0.005
TC214880 homologue to UP|P93169 (P93169) Early light-induced pro... 36 0.005
BE609443 36 0.007
BE800970 similar to SP|Q02060|PSBS_ Photosystem II 22 kDa protei... 35 0.012
TC204019 similar to GB|AAP21362.1|30102888|BT006554 At2g21970 {A... 35 0.012
TC220638 weakly similar to UP|Q7NKN4 (Q7NKN4) CAB/ELIP/HLIP-rela... 35 0.016
>TC204939 similar to GB|AAN38679.1|23507751|BT000842 At4g34190/F28A23_50
{Arabidopsis thaliana;} , partial (53%)
Length = 727
Score = 210 bits (535), Expect = 2e-55
Identities = 112/143 (78%), Positives = 121/143 (84%)
Frame = +3
Query: 1 MALATQTSASLSISFRDACVFPAAKYSATARFPLPNLVPSRASFATGTPLLIRRSHGRKL 60
MAL Q ASLSISFRDA VF A K S T FP NLVP+RASF G+PLLIRR+HGR
Sbjct: 105 MALV-QPFASLSISFRDARVFTAPKCSPTTHFPHRNLVPTRASFIAGSPLLIRRNHGRNG 281
Query: 61 ACRAMPVSIRSEQSTKDGNGLDVWLGRLAMIGFAAGITVEVATGKGLLENFGVASPLPTV 120
AC+AMPVSIR EQST++GN LDVWLGRLAM+GFA ITVE+ATGKGLLENFG+ SPLPTV
Sbjct: 282 ACKAMPVSIRCEQSTQEGNSLDVWLGRLAMVGFAVAITVEIATGKGLLENFGLTSPLPTV 461
Query: 121 ALAVTALVGVLTAVFIFQSASKN 143
ALAVTALVGVLTAVFIFQSASKN
Sbjct: 462 ALAVTALVGVLTAVFIFQSASKN 530
>TC207501 similar to GB|AAN38679.1|23507751|BT000842 At4g34190/F28A23_50
{Arabidopsis thaliana;} , partial (52%)
Length = 724
Score = 185 bits (469), Expect = 7e-48
Identities = 101/143 (70%), Positives = 115/143 (79%)
Frame = +3
Query: 1 MALATQTSASLSISFRDACVFPAAKYSATARFPLPNLVPSRASFATGTPLLIRRSHGRKL 60
MAL + ASLSIS A +F K S TA FP N V +R SF G+PLLI+R+HGRK
Sbjct: 93 MALV-EAYASLSIS-SVARLFTPPKCSPTAHFPHRNFVATRPSFIVGSPLLIQRNHGRKA 266
Query: 61 ACRAMPVSIRSEQSTKDGNGLDVWLGRLAMIGFAAGITVEVATGKGLLENFGVASPLPTV 120
AC+A+PV IR EQST+ GNGLDVWLGRLAM+GFA ITVE++TGKGLLENFGV +PLPTV
Sbjct: 267 ACKAIPVPIRCEQSTQQGNGLDVWLGRLAMVGFAVAITVEISTGKGLLENFGVPTPLPTV 446
Query: 121 ALAVTALVGVLTAVFIFQSASKN 143
ALAVTALVGVLTAVFIFQSASK+
Sbjct: 447 ALAVTALVGVLTAVFIFQSASKS 515
>TC204937 similar to GB|AAN38679.1|23507751|BT000842 At4g34190/F28A23_50
{Arabidopsis thaliana;} , partial (53%)
Length = 514
Score = 183 bits (465), Expect = 2e-47
Identities = 91/115 (79%), Positives = 102/115 (88%)
Frame = +3
Query: 29 TARFPLPNLVPSRASFATGTPLLIRRSHGRKLACRAMPVSIRSEQSTKDGNGLDVWLGRL 88
TA FP N V +R SF +G+PLLIRR+HGRK AC+AMPV+IR EQST++GN LDVWLGRL
Sbjct: 6 TAHFPHRNFVATRPSFISGSPLLIRRNHGRKAACKAMPVTIRCEQSTQEGNSLDVWLGRL 185
Query: 89 AMIGFAAGITVEVATGKGLLENFGVASPLPTVALAVTALVGVLTAVFIFQSASKN 143
AM+GFA ITVE+ATGKGLLENFG+ SPLPTVALAVTALVGVLTAVFIFQSASKN
Sbjct: 186 AMVGFAVAITVEIATGKGLLENFGLTSPLPTVALAVTALVGVLTAVFIFQSASKN 350
>TC204942 homologue to UP|Q6W5F3 (Q6W5F3) Microtubule-associated protein 1
light chain 3, partial (84%)
Length = 1000
Score = 142 bits (358), Expect = 5e-35
Identities = 72/90 (80%), Positives = 78/90 (86%)
Frame = -2
Query: 53 RRSHGRKLACRAMPVSIRSEQSTKDGNGLDVWLGRLAMIGFAAGITVEVATGKGLLENFG 112
R G + AC AMPVSIR E ST++GN LDVWLGRLAM+GFA ITVE+ATGKGLLENFG
Sbjct: 429 RAEFGTRAAC*AMPVSIRCEPSTQEGNSLDVWLGRLAMVGFAVAITVEIATGKGLLENFG 250
Query: 113 VASPLPTVALAVTALVGVLTAVFIFQSASK 142
+ SPLPTVALAVTALVGVLTAVFIFQSASK
Sbjct: 249 LTSPLPTVALAVTALVGVLTAVFIFQSASK 160
>AW459109
Length = 398
Score = 128 bits (321), Expect = 1e-30
Identities = 64/81 (79%), Positives = 71/81 (87%)
Frame = +1
Query: 63 RAMPVSIRSEQSTKDGNGLDVWLGRLAMIGFAAGITVEVATGKGLLENFGVASPLPTVAL 122
RAMPVSIR EQST++GN L+VWLGRLAM+GFA ITVE+ATGKGLLENF + SPLPT L
Sbjct: 4 RAMPVSIRCEQSTQEGNSLNVWLGRLAMVGFAVTITVEIATGKGLLENFKLTSPLPTXTL 183
Query: 123 AVTALVGVLTAVFIFQSASKN 143
VTALV +LTAVFIFQSASKN
Sbjct: 184 TVTALVDILTAVFIFQSASKN 246
>AW102413
Length = 364
Score = 117 bits (292), Expect = 2e-27
Identities = 65/121 (53%), Positives = 78/121 (63%)
Frame = +2
Query: 23 AAKYSATARFPLPNLVPSRASFATGTPLLIRRSHGRKLACRAMPVSIRSEQSTKDGNGLD 82
A K S T FP NLVP++ASF TG+PLLIR +H R AC AMPV IR EQST +GN LD
Sbjct: 2 APKCSPTTHFPHRNLVPTQASFITGSPLLIRTNHERNGACEAMPV*IRYEQSTHEGNSLD 181
Query: 83 VWLGRLAMIGFAAGITVEVATGKGLLENFGVASPLPTVALAVTALVGVLTAVFIFQSASK 142
+ L +LA++ F IT++ AT + LL NF + SPL T L T L LTA IFQ S
Sbjct: 182 IGLNQLAIVRFTLTITIKTATRRELLYNFTLTSPLATCTLT*TTLTNSLTASAIFQXPSH 361
Query: 143 N 143
N
Sbjct: 362 N 364
>TC204260 similar to UP|PSBS_SPIOL (Q02060) Photosystem II 22 kDa protein,
chloroplast precursor (CP22), partial (74%)
Length = 1225
Score = 42.7 bits (99), Expect = 6e-05
Identities = 36/135 (26%), Positives = 60/135 (43%), Gaps = 17/135 (12%)
Frame = +3
Query: 1 MALATQTSASLSISFRDACVFPAAKYSATARF------PLPNLVPSRASFATGTPLLIRR 54
M L + S+S S+ + S +F PLP+ S +S T T L + +
Sbjct: 111 MLLMSSVSSSYSVDLKKDLFLQLQSQSLRPKFSQLSFNPLPSSTSSFSSPRTFTTLALFK 290
Query: 55 SHGRKLACRAMPVSIRSEQSTKDG-----------NGLDVWLGRLAMIGFAAGITVEVAT 103
S + A A + +Q +DG ++++GR+AM+GFAA + E T
Sbjct: 291 SKTK--AAPAKTKVTKPKQKVEDGIFGTSGGFGFTKQNELFVGRVAMLGFAASLLGEGIT 464
Query: 104 GKGLLENFGVASPLP 118
GKG+L + + +P
Sbjct: 465 GKGILAQLNLETGIP 509
Score = 34.7 bits (78), Expect = 0.016
Identities = 16/50 (32%), Positives = 27/50 (54%)
Frame = +3
Query: 82 DVWLGRLAMIGFAAGITVEVATGKGLLENFGVASPLPTVALAVTALVGVL 131
++++GRLA +GF + E+ TGKG L + + +P + L VL
Sbjct: 702 ELFVGRLAQLGFVFSLIGEIITGKGALAQLNIETGVPINEIEPLVLFNVL 851
>BU550192 homologue to PIR|JC5876|JC5 early light-inducible protein precursor
- soybean, partial (76%)
Length = 657
Score = 39.7 bits (91), Expect = 5e-04
Identities = 28/61 (45%), Positives = 38/61 (61%), Gaps = 4/61 (6%)
Frame = -3
Query: 86 GRLAMIGFAAGITVEVATGKGLLE---NFGVASPLPT-VALAVTALVGVLTAVFIFQSAS 141
GRLAMIGF A + VEVA G+G+LE N G+ L T V L + +L+ + V + +S S
Sbjct: 364 GRLAMIGFVAAMAVEVAKGQGVLEQISNGGIPWFLGTSVVLTLASLIPLFQGVSV-ESKS 188
Query: 142 K 142
K
Sbjct: 187 K 185
Score = 25.4 bits (54), Expect = 9.7
Identities = 13/42 (30%), Positives = 20/42 (46%)
Frame = -3
Query: 67 VSIRSEQSTKDGNGLDVWLGRLAMIGFAAGITVEVATGKGLL 108
VS+ S+ + ++W GR AM+G A E G L+
Sbjct: 208 VSVESKSKGFMSSDAELWNGRFAMLGLIALAFTEYVKGSTLV 83
>TC204484 homologue to UP|P93169 (P93169) Early light-induced protein,
complete
Length = 893
Score = 39.7 bits (91), Expect = 5e-04
Identities = 28/61 (45%), Positives = 38/61 (61%), Gaps = 4/61 (6%)
Frame = +1
Query: 86 GRLAMIGFAAGITVEVATGKGLLE---NFGVASPLPT-VALAVTALVGVLTAVFIFQSAS 141
GRLAMIGF A + VEVA G+G+LE N G+ L T V L + +L+ + V + +S S
Sbjct: 472 GRLAMIGFVAAMAVEVAKGQGVLEQISNGGIPWFLGTSVVLTLASLIPLFQGVSV-ESKS 648
Query: 142 K 142
K
Sbjct: 649 K 651
Score = 25.4 bits (54), Expect = 9.7
Identities = 13/42 (30%), Positives = 20/42 (46%)
Frame = +1
Query: 67 VSIRSEQSTKDGNGLDVWLGRLAMIGFAAGITVEVATGKGLL 108
VS+ S+ + ++W GR AM+G A E G L+
Sbjct: 628 VSVESKSKGFMSSDAELWNGRFAMLGLIALAFTEYVKGSTLV 753
>BG156750 similar to SP|P54773|PSBS Photosystem II 22 kDa protein
chloroplast precursor (CP22). [Tomato] {Lycopersicon
esculentum}, partial (44%)
Length = 466
Score = 37.7 bits (86), Expect = 0.002
Identities = 32/126 (25%), Positives = 58/126 (45%), Gaps = 11/126 (8%)
Frame = +2
Query: 4 ATQTSASLSISFRDACVFPAAKYSATARFPLPNLVPSRASFATGTPLLIRRSHGRKLACR 63
A LS+ + + P +S + PL + PS +S T T L + +S+ + A
Sbjct: 5 AVDMKKDLSLQLQSQRLRPT--FSQLSFNPLASSTPSFSSPRTFTTLALFKSNTK--AAP 172
Query: 64 AMPVSIRSEQSTKDG-----------NGLDVWLGRLAMIGFAAGITVEVATGKGLLENFG 112
A + +Q +DG ++++GR+AM+G+AA + E T KG+L
Sbjct: 173 AKTKVTKPKQKVEDGIFGTSGRFGSTKQNELFVGRVAMLGYAASLLGERITAKGILSQLN 352
Query: 113 VASPLP 118
+ + +P
Sbjct: 353 LETGIP 370
>TC212014 similar to UP|PSBS_ARATH (Q9XF91) Photosystem II 22 kDa protein,
chloroplast precursor (CP22), partial (37%)
Length = 829
Score = 37.0 bits (84), Expect = 0.003
Identities = 32/114 (28%), Positives = 56/114 (49%), Gaps = 1/114 (0%)
Frame = +2
Query: 22 PAAKYSATARFPLPNLVPSRASFATGTPLLIRRSHGRKLACRAMPVSIRSEQSTKDGNGL 81
P+A YS ++ + + A F T TP + +S + A+ S TK
Sbjct: 152 PSASYSFSSSSSR-TFITALAFFKTKTPSKVVKSKPKPKVEDAIFGSSGGFGFTKVN--- 319
Query: 82 DVWLGRLAMIGFAAGITVEVATGKGLLENFGVASPLPTV-ALAVTALVGVLTAV 134
++++GR+AM+GF+A E TGKG++ + +P AL + L+G + A+
Sbjct: 320 ELFVGRVAMLGFSAPSLGEAITGKGIIAQLNLEPGMPIYEALLLFTLLGAIGAL 481
>TC214884 similar to UP|P93169 (P93169) Early light-induced protein, partial
(98%)
Length = 2021
Score = 37.0 bits (84), Expect = 0.003
Identities = 17/24 (70%), Positives = 20/24 (82%)
Frame = +3
Query: 86 GRLAMIGFAAGITVEVATGKGLLE 109
GRLAMIGF A + VEVA G+G+LE
Sbjct: 1743 GRLAMIGFVAAMAVEVAKGQGVLE 1814
>BF068887 homologue to PIR|JC5876|JC58 early light-inducible protein
precursor - soybean, partial (48%)
Length = 413
Score = 37.0 bits (84), Expect = 0.003
Identities = 17/24 (70%), Positives = 20/24 (82%)
Frame = +3
Query: 86 GRLAMIGFAAGITVEVATGKGLLE 109
GRLAMIGF A + VEVA G+G+LE
Sbjct: 282 GRLAMIGFVAAMAVEVAKGQGVLE 353
>TC204259 similar to UP|PSBS_SPIOL (Q02060) Photosystem II 22 kDa protein,
chloroplast precursor (CP22), partial (72%)
Length = 1043
Score = 36.2 bits (82), Expect = 0.005
Identities = 15/37 (40%), Positives = 26/37 (69%)
Frame = +3
Query: 82 DVWLGRLAMIGFAAGITVEVATGKGLLENFGVASPLP 118
++++GR+AM+GFAA + E TGKG+L + + +P
Sbjct: 339 ELFVGRVAMLGFAASLLGEGITGKGILAQLNLETGIP 449
Score = 34.7 bits (78), Expect = 0.016
Identities = 16/50 (32%), Positives = 27/50 (54%)
Frame = +3
Query: 82 DVWLGRLAMIGFAAGITVEVATGKGLLENFGVASPLPTVALAVTALVGVL 131
++++GRLA +GF + E+ TGKG L + + +P + L VL
Sbjct: 642 ELFVGRLAQLGFVFSLIGEIITGKGALAQLNIETGVPINEIEPLVLFNVL 791
>TC214885 homologue to UP|P93169 (P93169) Early light-induced protein,
complete
Length = 1256
Score = 36.2 bits (82), Expect = 0.005
Identities = 25/61 (40%), Positives = 36/61 (58%), Gaps = 4/61 (6%)
Frame = +1
Query: 86 GRLAMIGFAAGITVEVATGKGLLE---NFGVASPLPT-VALAVTALVGVLTAVFIFQSAS 141
GRLAMIGF + VEVA G+G+ E N G+ L T V L + +L+ + + + +S S
Sbjct: 853 GRLAMIGFVGAMAVEVAKGQGVFEQISNGGIPWFLGTSVVLTLASLIPLFQGISV-ESKS 1029
Query: 142 K 142
K
Sbjct: 1030K 1032
>TC214880 homologue to UP|P93169 (P93169) Early light-induced protein,
complete
Length = 940
Score = 36.2 bits (82), Expect = 0.005
Identities = 25/61 (40%), Positives = 36/61 (58%), Gaps = 4/61 (6%)
Frame = +3
Query: 86 GRLAMIGFAAGITVEVATGKGLLE---NFGVASPLPT-VALAVTALVGVLTAVFIFQSAS 141
GRLAMIGF + VEVA G+G+ E N G+ L T V L + +L+ + + + +S S
Sbjct: 432 GRLAMIGFVGAMAVEVAKGQGVFEQISNGGIPWFLGTSVVLTLASLIPLFQGISV-ESKS 608
Query: 142 K 142
K
Sbjct: 609 K 611
>BE609443
Length = 458
Score = 35.8 bits (81), Expect = 0.007
Identities = 15/37 (40%), Positives = 26/37 (69%)
Frame = +1
Query: 82 DVWLGRLAMIGFAAGITVEVATGKGLLENFGVASPLP 118
++++GR+AM+GFAA + E TGKG+L + + +P
Sbjct: 124 ELFVGRVAMLGFAASLLGEGNTGKGILAQLNLETGIP 234
>BE800970 similar to SP|Q02060|PSBS_ Photosystem II 22 kDa protein
chloroplast precursor (CP22). [Spinach] {Spinacia
oleracea}, partial (21%)
Length = 181
Score = 35.0 bits (79), Expect = 0.012
Identities = 17/48 (35%), Positives = 28/48 (57%)
Frame = +2
Query: 84 WLGRLAMIGFAAGITVEVATGKGLLENFGVASPLPTVALAVTALVGVL 131
++GR+AM+GFA + E TGKG+L + + + +P L G+L
Sbjct: 2 FVGRVAMLGFAESLLGEGITGKGILAHLNLQTGMPINEAKPHLLYGIL 145
>TC204019 similar to GB|AAP21362.1|30102888|BT006554 At2g21970 {Arabidopsis
thaliana;} , partial (75%)
Length = 922
Score = 35.0 bits (79), Expect = 0.012
Identities = 19/71 (26%), Positives = 38/71 (52%)
Frame = +3
Query: 73 QSTKDGNGLDVWLGRLAMIGFAAGITVEVATGKGLLENFGVASPLPTVALAVTALVGVLT 132
+S+K + ++ GRLAM+ FAA +T+E+ TG + + + + A+
Sbjct: 321 ESSKKSHDFEIISGRLAMMVFAATVTMEMVTGNSMFRKMDIEGITEAGGVCLGAV--TCA 494
Query: 133 AVFIFQSASKN 143
A+F + S+++N
Sbjct: 495 ALFAWFSSARN 527
>TC220638 weakly similar to UP|Q7NKN4 (Q7NKN4) CAB/ELIP/HLIP-related protein,
partial (62%)
Length = 781
Score = 34.7 bits (78), Expect = 0.016
Identities = 19/51 (37%), Positives = 29/51 (56%)
Frame = +1
Query: 86 GRLAMIGFAAGITVEVATGKGLLENFGVASPLPTVALAVTALVGVLTAVFI 136
GR+A++G AA + VE+ATGKG++ P + L V + AVF+
Sbjct: 370 GRIAILGLAALVLVELATGKGVINYH-----TPAIILIQIYFVAAVGAVFV 507
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.133 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,162,780
Number of Sequences: 63676
Number of extensions: 60196
Number of successful extensions: 449
Number of sequences better than 10.0: 113
Number of HSP's better than 10.0 without gapping: 438
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 449
length of query: 143
length of database: 12,639,632
effective HSP length: 88
effective length of query: 55
effective length of database: 7,036,144
effective search space: 386987920
effective search space used: 386987920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0290.12


