

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0283.14
(461 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
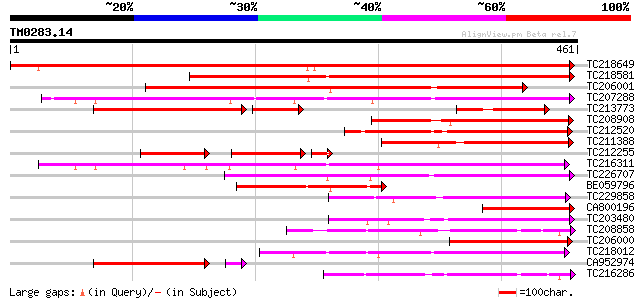
Score E
Sequences producing significant alignments: (bits) Value
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 568 e-162
TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F2... 421 e-118
TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, parti... 257 7e-69
TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete 163 2e-40
TC213773 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, parti... 115 4e-35
TC208908 weakly similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-... 139 3e-33
TC212520 137 1e-32
TC211388 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like pr... 132 2e-31
TC212255 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F2... 79 6e-31
TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete 128 6e-30
TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 117 1e-26
BE059796 112 5e-25
TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporte... 107 9e-24
CA800196 107 1e-23
TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, par... 106 2e-23
TC208858 similar to UP|Q9M9Z8 (Q9M9Z8) F20B17.24, partial (51%) 106 2e-23
TC206000 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like pr... 103 2e-22
TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporte... 101 8e-22
CA952974 similar to GP|17381265|gb| AT5g18840/F17K4_90 {Arabidop... 98 1e-21
TC216286 similar to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partia... 100 2e-21
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 568 bits (1465), Expect = e-162
Identities = 295/485 (60%), Positives = 363/485 (74%), Gaps = 26/485 (5%)
Frame = +2
Query: 1 MPLGEDYEDSRNIRKPFINNNN-----------------AGSGSFFVVLCVLVVALGPIQ 43
M L ED E+ R+++KPF++ + S V CVL+VALGPIQ
Sbjct: 35 MSLREDNEEGRDLKKPFLHTGSWYRMSGRQSSVFGSTQAIRDSSISVFACVLIVALGPIQ 214
Query: 44 FGFTCGYSSPTQEEMIRDLNLSISRFSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVA 103
FGFT GY+SPTQ +I DL LS+S FSLFGSLSNVGAMVGA SG +AEY GRKGSL++A
Sbjct: 215 FGFTAGYTSPTQSAIINDLGLSVSEFSLFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIA 394
Query: 104 AVPNIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQL 163
++PNI GW+AIS AKDSS L++GRLLEGFGVGIISY VPVYIAEI P +RG L +VNQL
Sbjct: 395 SIPNIIGWLAISFAKDSSFLYMGRLLEGFGVGIISYTVPVYIAEISPPNLRGGLVSVNQL 574
Query: 164 SVTIGIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQ 223
SVTIGIMLAYLLG+F WR LA++G+ PC ILIP L+FIPESPRWLA+MG+ E FE+SLQ
Sbjct: 575 SVTIGIMLAYLLGIFVEWRILAIIGILPCTILIPALFFIPESPRWLAKMGMTEEFETSLQ 754
Query: 224 TLRGSNVDITMEAQEIQ------NRFYTV---CRPQEEKILVSFDGIGLLVLQQLSGING 274
LRG + DI++E EI+ N TV Q L GIGLL+LQQLSGING
Sbjct: 755 VLRGFDTDISVEVNEIKRAVASTNTRITVRFADLKQRRYWLPLMIGIGLLILQQLSGING 934
Query: 275 VFFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLL 334
V FY+S IF +AGISSSDAATFG+GA+QV+ T + WL DKSGRR+LLIVS++ M+ SLL
Sbjct: 935 VLFYSSTIFRNAGISSSDAATFGVGAVQVLATSLTLWLADKSGRRLLLIVSATGMSFSLL 1114
Query: 335 LVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGL 394
+VA FY++ +S+ S L+ IL LS+VG+VA++I F+LG+G +PW+IMSEILP NIKGL
Sbjct: 1115VVAITFYIKASISETSSLYGILSTLSLVGVVAMVIAFSLGMGAMPWIIMSEILPINIKGL 1294
Query: 395 AGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEE 454
AGSVAT +NW + ++TLTAN LL+WSS GTFTIYA+ A TV F ++VPETK +T+EE
Sbjct: 1295AGSVATLANWLFSWLVTLTANMLLDWSSGGTFTIYAVVCALTVVFVTIWVPETKGKTIEE 1474
Query: 455 IQASF 459
IQ SF
Sbjct: 1475IQWSF 1489
>TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;} , partial (66%)
Length = 1340
Score = 421 bits (1082), Expect = e-118
Identities = 213/324 (65%), Positives = 263/324 (80%), Gaps = 11/324 (3%)
Frame = +2
Query: 147 EIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESP 206
EI P + G LG+VNQLS+TIGIMLAYLLGLF NWR LA+LG+ PC +LIPGL+FIPESP
Sbjct: 5 EIAPNHLIGGLGSVNQLSITIGIMLAYLLGLFVNWRILAILGILPCTVLIPGLFFIPESP 184
Query: 207 RWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQN-----------RFYTVCRPQEEKIL 255
RWLA+MG+++ FE+SLQ LRG + DI++E EI+ RF + R + L
Sbjct: 185 RWLAKMGMIDEFETSLQVLRGFDTDISVEVHEIKRSVASTGKRAAIRFADLKRKRYWFPL 364
Query: 256 VSFDGIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDK 315
+ GIGLLVLQQLSGING+ FY++ IFA+AGISSS+AAT GLGA+QV+ TG++TWLVDK
Sbjct: 365 MV--GIGLLVLQQLSGINGILFYSTTIFANAGISSSEAATVGLGAVQVIATGISTWLVDK 538
Query: 316 SGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGI 375
SGRR+LLI+SSS+MT+SLL+V+ AFYLEGVVS+ S L +ILGI+S+VGLVA++IGF+LG+
Sbjct: 539 SGRRLLLIISSSVMTVSLLIVSIAFYLEGVVSEDSHLFSILGIVSIVGLVAMVIGFSLGL 718
Query: 376 GPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAF 435
GPIPWLIMSEILP NIKGLAGS+AT NW + IT+TAN LLNWSS GTFTIY + +AF
Sbjct: 719 GPIPWLIMSEILPVNIKGLAGSIATMGNWLISWGITMTANLLLNWSSGGTFTIYTVVAAF 898
Query: 436 TVAFALLFVPETKDRTLEEIQASF 459
T+AF ++VPETK RTLEEIQ SF
Sbjct: 899 TIAFIAMWVPETKGRTLEEIQFSF 970
>TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, partial (66%)
Length = 951
Score = 257 bits (657), Expect = 7e-69
Identities = 138/320 (43%), Positives = 198/320 (61%), Gaps = 9/320 (2%)
Frame = +3
Query: 111 WVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIM 170
W+A+ +K S L +GR G+G+G+ISYVVPVYIAEI P+ +RG L NQL + G
Sbjct: 3 WLAVFFSKGSYSLDMGRFFTGYGIGVISYVVPVYIAEIAPKNLRGGLATTNQLLIVTGGS 182
Query: 171 LAYLLGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNV 230
+++LLG NWR LAL G+ PC L+ GL FIPESPRWLA++G + F+ +L LRG +
Sbjct: 183 VSFLLGSVINWRELALAGLVPCICLLVGLCFIPESPRWLAKVGREKEFQLALSRLRGKHA 362
Query: 231 DITMEAQEIQNRFYTVCRPQEEKILVSFD---------GIGLLVLQQLSGINGVFFYASK 281
DI+ EA EI + T+ + K+L F G+GL+ QQ GING+ FY ++
Sbjct: 363 DISDEAAEILDYIETLESLPKTKLLDLFQSKYVHSVVIGVGLMACQQSVGINGIGFYTAE 542
Query: 282 IFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFY 341
IF +AG+SS A T IQ+ T + L+DKSGRR L++VS++ + + A AF+
Sbjct: 543 IFVAAGLSSGKAGTIAYACIQIPFTLLGAILMDKSGRRPLVMVSAAGTFLGCFVAAFAFF 722
Query: 342 LEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATF 401
L+ S L + IL+ G++ I F++G+G +PW+IMSEI P ++KG AGS+
Sbjct: 723 LK----DQSLLPEWVPILAFAGVLIYIAAFSIGLGSVPWVIMSEIFPIHLKGTAGSLVVL 890
Query: 402 SNWFTASVITLTANFLLNWS 421
W A V++ T NFL++WS
Sbjct: 891 VAWLGAWVVSYTFNFLMSWS 950
>TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete
Length = 1813
Score = 163 bits (412), Expect = 2e-40
Identities = 127/471 (26%), Positives = 223/471 (46%), Gaps = 38/471 (8%)
Frame = +1
Query: 27 SFFVVLCVLVVALGPIQFGFTCGYSS--PTQEEMIRDLNLSISR---------------- 68
++F CV V ALG FG+ G S P+ ++ +++ + R
Sbjct: 97 AYFAFTCV-VGALGGSLFGYDLGVSGGVPSMDDFLKEFFPKVYRRKQMHLHETDYCKYDD 273
Query: 69 --FSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIG 126
+LF S A+V + L GRK S++V A+ + G + + AK+ ++L IG
Sbjct: 274 QVLTLFTSSLYFSALVMTFFASFLTRKKGRKASIIVGALSFLAGAILNAAAKNIAMLIIG 453
Query: 127 RLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLF------FN 180
R+L G G+G + VP+Y++E+ P RG++ + Q + GI++A L+ F +
Sbjct: 454 RVLLGGGIGFGNQAVPLYLSEMAPAKNRGAVNQLFQFTTCAGILIANLVNYFTEKIHPYG 633
Query: 181 WR-TLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGS-NV-------- 230
WR +L L G+ A+L+ G+ E+P L E G +++ + LQ +RG+ NV
Sbjct: 634 WRISLGLAGLPAFAMLVGGI-CCAETPNSLVEQGRLDKAKQVLQRIRGTENVEAEFEDLK 810
Query: 231 DITMEAQEIQNRFYTVCRPQEEKILVSFDGIGLLVLQQLSGINGVFFYASKIFASAGISS 290
+ + EAQ +++ F T+ + + L+ +G+ QQL+G N + FYA IF S G +
Sbjct: 811 EASEEAQAVKSPFRTLLKRKYRPQLI-IGALGIPAFQQLTGNNSILFYAPVIFQSLGFGA 987
Query: 291 SDA--ATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSK 348
+ + ++F +V T ++ +LVDK GRR + + M +++ + +
Sbjct: 988 NASLFSSFITNGALLVATVISMFLVDKYGRRKFFLEAGFEMICCMIITGAVLAVN--FGH 1161
Query: 349 GSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTAS 408
G E+ + VV + ++ + GP+ WL+ SE+ P I+ A S+ N +
Sbjct: 1162GKEIGKGVSAFLVVVIFLFVLAYGRSWGPLGWLVPSELFPLEIRSSAQSIVVCVNMIFTA 1341
Query: 409 VITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQASF 459
++ L G F ++A F F +PETK +EEI F
Sbjct: 1342LVAQLFLMSLCHLKFGIFLLFASLIIFMSFFVFFLLPETKKVPIEEIYLLF 1494
>TC213773 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, partial (49%)
Length = 730
Score = 115 bits (287), Expect(2) = 4e-35
Identities = 56/124 (45%), Positives = 79/124 (63%)
Frame = +3
Query: 69 FSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRL 128
FS+FGSL +GAM+GA SG + ++ GRKG++ ++A I G + +K S L +GR
Sbjct: 12 FSMFGSLVTIGAMLGAISSGRITDFIGRKGAMRISAGFCITGCFFVFFSKGSYSLDLGRF 191
Query: 129 LEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQLSVTIGIMLAYLLGLFFNWRTLALLG 188
G+G+G+ISYVVPVYI EI P+ +R L NQL + +++LLG NWR LAL G
Sbjct: 192 FTGYGIGVISYVVPVYIVEIAPKNLREELATTNQLLIVTEASVSFLLGSVINWRKLALAG 371
Query: 189 VFPC 192
+ C
Sbjct: 372 LVSC 383
Score = 65.9 bits (159), Expect = 4e-11
Identities = 31/76 (40%), Positives = 47/76 (61%)
Frame = +2
Query: 364 LVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSA 423
++ I +++G GP+PW+IMSE G+AGS+ NW A +++ T NFL++WSS
Sbjct: 521 ILIYIAAYSIGEGPVPWVIMSE-------GIAGSLVVLVNWLGAWIVSYTFNFLMSWSSP 679
Query: 424 GTFTIYAIFSAFTVAF 439
GT +YA S T+ F
Sbjct: 680 GT*FLYAGSSLLTILF 727
Score = 51.6 bits (122), Expect(2) = 4e-35
Identities = 25/42 (59%), Positives = 33/42 (78%)
Frame = +2
Query: 198 GLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEI 239
GL FIPESPRWLA++G + F+ +L+ LRG +VDI+ EA EI
Sbjct: 398 GLCFIPESPRWLAKVGREK*FQLALRRLRGKDVDISDEAAEI 523
>TC208908 weakly similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like
protein, partial (28%)
Length = 753
Score = 139 bits (350), Expect = 3e-33
Identities = 74/167 (44%), Positives = 108/167 (64%), Gaps = 3/167 (1%)
Frame = +2
Query: 295 TFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHN 354
T + A+++ MT + L+DKSGRR LL+VS+ + L A +F L+ +LH
Sbjct: 2 TIAIVAVKIPMTTIGVLLMDKSGRRPLLLVSAVGTCVGCFLAALSFVLQ-------DLHK 160
Query: 355 ILG---ILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVIT 411
G IL++VG++ + +++G+G IPW+IMSEI P N+KG AGS+ T +W + +I+
Sbjct: 161 WKGVSPILALVGVLVYVGSYSIGMGAIPWVIMSEIFPINVKGSAGSLVTLVSWLCSWIIS 340
Query: 412 LTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQAS 458
+ NFL++WSSAGTF +++ FTV F VPETK RTLEEIQAS
Sbjct: 341 YSFNFLMSWSSAGTFLMFSSICGFTVLFVAKLVPETKGRTLEEIQAS 481
>TC212520
Length = 720
Score = 137 bits (345), Expect = 1e-32
Identities = 76/185 (41%), Positives = 112/185 (60%)
Frame = +3
Query: 273 NGVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTIS 332
NG+ F + S S T +Q+V G+ L+DK+GR+ L+++S S +
Sbjct: 3 NGICFIPAVXXTSR--ISPTIGTITYACLQIVXXGLGAALIDKAGRKPLILLSGSGLVAG 176
Query: 333 LLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIK 392
VA AFYL+ V G E + L+V G++ I F++G+G IPW++M EI P NIK
Sbjct: 177 CTFVAVAFYLK-VHEVGVEA---VPALAVTGILVYIGSFSIGMGAIPWVVMCEIFPVNIK 344
Query: 393 GLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTL 452
GLAGSVAT NWF A + + T NF ++WSS GTF +YA +A + F ++ VPE+K ++L
Sbjct: 345 GLAGSVATLVNWFGAWLCSYTFNFFMSWSSYGTFILYAAINALDILFIIVAVPESKGKSL 524
Query: 453 EEIQA 457
E++QA
Sbjct: 525 EQLQA 539
>TC211388 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like protein,
partial (27%)
Length = 786
Score = 132 bits (333), Expect = 2e-31
Identities = 70/158 (44%), Positives = 100/158 (62%), Gaps = 2/158 (1%)
Frame = +3
Query: 303 VVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVS--KGSELHNILGILS 360
+ MT + L+DKSGRR LL++S+S + L A +F L+ + +GS + + G+L
Sbjct: 87 IPMTALGVLLMDKSGRRPLLLISASGTCLGCFLAALSFTLQDLHKWKEGSPILALAGVLV 266
Query: 361 VVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNW 420
G F+LG+G IPW+IMSEI P N+KG AGS+ T +W + +++ NFL++W
Sbjct: 267 YTG------SFSLGMGGIPWVIMSEIFPTNVKGSAGSLVTLVSWLCSWIVSYAFNFLMSW 428
Query: 421 SSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQAS 458
SSAGTF I++ FT+ F VPETK RTLEE+QAS
Sbjct: 429 SSAGTFFIFSSICGFTILFVAKLVPETKGRTLEEVQAS 542
>TC212255 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;} , partial (27%)
Length = 894
Score = 78.6 bits (192), Expect(3) = 6e-31
Identities = 33/60 (55%), Positives = 48/60 (80%)
Frame = +2
Query: 181 WRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQ 240
W ++ G+FPC +LI GL+FIP+SPRWLA+MG+++ FE+SLQ LRG + DI++E EI+
Sbjct: 452 WPWISEPGIFPCTVLIRGLFFIPKSPRWLAKMGMIDEFETSLQMLRGFDTDISVEVHEIK 631
Score = 73.9 bits (180), Expect(3) = 6e-31
Identities = 37/56 (66%), Positives = 44/56 (78%)
Frame = +3
Query: 107 NIFGWVAISLAKDSSLLFIGRLLEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQ 162
N FG + +DSS L++GRLLEGFGVGIISYVV VYIAEI P+ +RG LG+VNQ
Sbjct: 306 NPFGSTC*VVVQDSSFLYMGRLLEGFGVGIISYVVLVYIAEIAPQNLRGGLGSVNQ 473
Score = 36.6 bits (83), Expect = 0.024
Identities = 14/18 (77%), Positives = 17/18 (93%)
Frame = +1
Query: 369 IGFALGIGPIPWLIMSEI 386
IGF+LG+GPIPWLI SE+
Sbjct: 694 IGFSLGLGPIPWLITSEV 747
Score = 20.4 bits (41), Expect(3) = 6e-31
Identities = 10/17 (58%), Positives = 12/17 (69%)
Frame = +1
Query: 246 VCRPQEEKILVSFDGIG 262
+CR QEEKI S G+G
Sbjct: 670 ICRSQEEKIGFSL-GLG 717
>TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete
Length = 1925
Score = 128 bits (321), Expect = 6e-30
Identities = 123/474 (25%), Positives = 207/474 (42%), Gaps = 42/474 (8%)
Frame = +1
Query: 24 GSGSFFVVLCVLVVALGPIQFGFTCGYSS--PTQEEMIRDLNLSISR------------- 68
GS + FV + +V A+G + FG+ G S + + + S+ R
Sbjct: 145 GSLTPFVTVTCIVAAMGGLIFGYDIGISGGVTSMDPFLLKFFPSVFRKKNSDKTVNQYCQ 324
Query: 69 -----FSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLL 123
++F S + A++ + V+ + FGRK S+L + + G + A+ +L
Sbjct: 325 YDSQTLTMFTSSLYLAALLSSLVASTVTRRFGRKLSMLFGGLLFLVGALINGFAQHVWML 504
Query: 124 FIGRLLEGFGVGIISYV---VPVYIAEIPPRTMRGSLG--AVNQLSVTIGIMLAYLLGL- 177
+GR+L GFG+G + V +P+ I G+L +Q + G + LL L
Sbjct: 505 IVGRILLGFGIGFANQVCATLPI*NGFIQI*RSIGTLAFNCQSQFGIPRGQCVELLLWLK 684
Query: 178 ---FFNWRTLALLGVF-PCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRG-SNVD- 231
+ W+ G P I+ G +P++P + E G E+ ++ LQ +RG NVD
Sbjct: 685 SMVAWGWKIEVWEGAMVPALIITVGSLVLPDTPNSMIERGDREKAKAQLQRIRGIDNVDE 864
Query: 232 -------ITMEAQEIQNRFYTVCRPQEEKILVSFDGIGLLVLQQLSGINGVFFYASKIFA 284
+ + ++++ + + + + L + + QQL+GIN + FYA +F+
Sbjct: 865 EFNDLVAASESSSQVEHPWRNLLQRKYRPHLTM--AVLIPFFQQLTGINVIMFYAPVLFS 1038
Query: 285 SAGISSSDAATFGL--GAIQVVMTGVATWLVDKSGRRILLIVSSSIMTISLLLVASAFYL 342
S G A + G + VV T V+ + VDK GRR L + M I +VA+A
Sbjct: 1039SIGFKDDAALMSAVITGVVNVVATCVSIYGVDKWGRRALFLEGGVQMLICQAVVAAAIGA 1218
Query: 343 E-GVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATF 401
+ G +L I+ V+ + + FA GP+ WL+ SEI P I+ A S+
Sbjct: 1219KFGTDGNPGDLPKWYAIVVVLFICIYVSAFAWSWGPLGWLVPSEIFPLEIRSAAQSINVS 1398
Query: 402 SNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEI 455
N +I +L G F +A F F F+PETK +EE+
Sbjct: 1399VNMLFTFLIAQVFLTMLCHMKFGLFLFFAFFVLIMTFFVYFFLPETKGIPIEEM 1560
>TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 2338
Score = 117 bits (292), Expect = 1e-26
Identities = 86/313 (27%), Positives = 145/313 (45%), Gaps = 28/313 (8%)
Frame = +3
Query: 175 LGLFFNWRTLALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITM 234
L L WR + +G P ++ + +PESPRWL G + + L + S + +
Sbjct: 42 LALRLGWRLMLGVGAIPSILIGVAVLAMPESPRWLVAKGRLGEAKRVLYKISESEEEARL 221
Query: 235 EAQEIQNRFYTVCRPQEEKILVS------------------------FDGIGLLVLQQLS 270
+I++ ++ +LVS +G+ Q +
Sbjct: 222 RLADIKDTAGIPQDCDDDVVLVSKQTHGHGVWRELFLHPTPAVRHIFIASLGIHFFAQAT 401
Query: 271 GINGVFFYASKIFASAGISSSD---AATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSS 327
GI+ V Y+ +IF AGI S + AT +G ++ V VAT+ +D++GRR+LL+ S S
Sbjct: 402 GIDAVVLYSPRIFEKAGIKSDNYRLLATVAVGFVKTVSILVATFFLDRAGRRVLLLCSVS 581
Query: 328 IMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEIL 387
+ +SLL + + VV N LS+ +++ + F++G GPI W+ SEI
Sbjct: 582 GLILSLLTLGLSLT---VVDHSQTTLNWAVGLSIAAVLSYVATFSIGSGPITWVYSSEIF 752
Query: 388 PPNIKGLAGSVATFSNWFTASVITLTANFLLN-WSSAGTFTIYAIFSAFTVAFALLFVPE 446
P ++ ++ N T+ VI +T L + G F ++A +A F +PE
Sbjct: 753 PLRLRAQGVAIGAAVNRVTSGVIAMTFLSLQKAITIGGAFFLFAGVAAVAWIFHYTLLPE 932
Query: 447 TKDRTLEEIQASF 459
T+ +TLEEI+ SF
Sbjct: 933 TRGKTLEEIEKSF 971
>BE059796
Length = 410
Score = 112 bits (279), Expect = 5e-25
Identities = 62/131 (47%), Positives = 82/131 (62%), Gaps = 9/131 (6%)
Frame = +1
Query: 185 ALLGVFPCAILIPGLYFIPESPRWLAEMGLMERFESSLQTLRGSNVDITMEAQEIQNRFY 244
ALLG+ PC + + GL+FIPESPRWLA+ G ER ES LQ LRG N D++ EA EI++ F
Sbjct: 1 ALLGIIPCIVQLLGLFFIPESPRWLAKFGHWERSESVLQRLRGKNADVSQEATEIRD-FT 177
Query: 245 TVCRPQEEKILVSFD---------GIGLLVLQQLSGINGVFFYASKIFASAGISSSDAAT 295
+ + E I+ F G+GL++LQQ G+NG+ FYAS IF SAG S S
Sbjct: 178 EALQRETESIIGLFQLQYLKSLTVGVGLMILQQFGGVNGIAFYASSIFISAGFSGS-IGM 354
Query: 296 FGLGAIQVVMT 306
+ A+QV+ T
Sbjct: 355 ITMVAVQVLHT 387
>TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporter 10 (Mus
musculus 13 days embryo heart cDNA, RIKEN full-length
enriched library, clone:D330001H08 product:solute
carrier family 2 (facilitated glucose transporter),
member 10, full insert sequence), partial (4%)
Length = 715
Score = 107 bits (268), Expect = 9e-24
Identities = 66/202 (32%), Positives = 106/202 (51%), Gaps = 5/202 (2%)
Frame = +3
Query: 260 GIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVAT----WLVDK 315
G GLL QQ +GIN V +Y+ I AG +++ A L I M T +L+D
Sbjct: 27 GAGLLAFQQFTGINTVMYYSPTIVQMAGFHANELALL-LSLIVAGMNAAGTILGIYLIDH 203
Query: 316 SGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGI 375
+GR+ L + S + +SL+++A AFY + S + + G L+VVGL I F+ G+
Sbjct: 204 AGRKKLALSSLGGVIVSLVILAFAFYKQSSTS-----NELYGWLAVVGLALYIGFFSPGM 368
Query: 376 GPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAG-TFTIYAIFSA 434
GP+PW + SEI P +G+ G ++ W + +++ T + G TF I + +
Sbjct: 369 GPVPWTLSSEIYPEEYRGICGGMSATVCWVSNLIVSETFLSIAEGIGIGSTFLIIGVIAV 548
Query: 435 FTVAFALLFVPETKDRTLEEIQ 456
F L++VPETK T +E++
Sbjct: 549 VAFVFVLVYVPETKGLTFDEVE 614
>CA800196
Length = 431
Score = 107 bits (267), Expect = 1e-23
Identities = 51/75 (68%), Positives = 60/75 (80%)
Frame = -1
Query: 385 EILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFV 444
+ILP NIKGLAGS+AT NW + IT+TAN LLNWSS GTFTIY + +AFT+AF ++V
Sbjct: 389 QILPVNIKGLAGSIATMGNWLISWGITMTANLLLNWSSGGTFTIYTVVAAFTIAFIAMWV 210
Query: 445 PETKDRTLEEIQASF 459
PETK RTLEEIQ SF
Sbjct: 209 PETKGRTLEEIQFSF 165
>TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (38%)
Length = 1040
Score = 106 bits (265), Expect = 2e-23
Identities = 70/213 (32%), Positives = 109/213 (50%), Gaps = 13/213 (6%)
Frame = +2
Query: 260 GIGLLVLQQLSGINGVFFYASKIFASAGIS--------SSDAATFGLGAIQVVMT----G 307
G+G+ +LQQ SGINGV +Y +I AG+ S++A+F + A + G
Sbjct: 218 GVGIQILQQFSGINGVLYYTPQILEEAGVEVLLSDIGIGSESASFLISAFTTFLMLPCIG 397
Query: 308 VATWLVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVAL 367
VA L+D SGRR LL+ + ++ +SL+++ + +V+ G+ H +S V +V
Sbjct: 398 VAMKLMDVSGRRQLLLTTIPVLIVSLIILV----IGSLVNFGNVAH---AAISTVCVVVY 556
Query: 368 IIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLL-NWSSAGTF 426
F +G GPIP ++ SEI P ++GL ++ W +IT + +L + G F
Sbjct: 557 FCCFVMGYGPIPNILCSEIFPTRVRGLCIAICALVFWIGDIIITYSLPVMLGSLGLGGVF 736
Query: 427 TIYAIFSAFTVAFALLFVPETKDRTLEEIQASF 459
IYA+ + F L VPETK LE I F
Sbjct: 737 AIYAVVCFISWIFVFLKVPETKGMPLEVISEFF 835
>TC208858 similar to UP|Q9M9Z8 (Q9M9Z8) F20B17.24, partial (51%)
Length = 993
Score = 106 bits (265), Expect = 2e-23
Identities = 82/241 (34%), Positives = 125/241 (51%), Gaps = 6/241 (2%)
Frame = +3
Query: 226 RGSNVDITMEAQEIQNRFYTVCRPQEEKILVSFDGIGLLVLQQLSGINGVFFYASKIFAS 285
RG D ++ I R++ V F G L LQQLSGIN VF+++S +F S
Sbjct: 87 RGDGSDSVKLSELIYGRYFRVM----------FIGSTLFALQQLSGINAVFYFSSTVFES 236
Query: 286 AGISSSDAATFGLGAIQVVMTGVATWLVDKSGRRILLIVSSSIMTIS--LLLVASAFYLE 343
G+ S D A +G ++ + VA L+DK GR++LL+ S M +S L ++A++ +
Sbjct: 237 FGVPS-DIANSCVGVCNLLGSVVAMILMDKLGRKVLLLGSFLGMGLSMGLQVIAASSFAS 413
Query: 344 GVVSKGSELHNILGILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSN 403
G S LSV G++ ++ FA G GP+P LIMSEILP NI+ A ++ +
Sbjct: 414 GFGSM---------YLSVGGMLLFVLSFAFGAGPVPCLIMSEILPSNIRAKAMAICLAVH 566
Query: 404 WFTASVITLTANFLLNWSSAGTFTIYAIFSAFTVAFALLFVP----ETKDRTLEEIQASF 459
W + L FL G +Y+IF F A++FV ETK ++L+EI+ +
Sbjct: 567 WVINFFVGLF--FLRLLELIGAQLLYSIF-GFCCLIAVVFVKKNILETKGKSLQEIEIAL 737
Query: 460 I 460
+
Sbjct: 738 L 740
>TC206000 similar to UP|Q8LBI9 (Q8LBI9) Sugar transporter-like protein,
partial (22%)
Length = 602
Score = 103 bits (256), Expect = 2e-22
Identities = 49/100 (49%), Positives = 67/100 (67%)
Frame = +1
Query: 358 ILSVVGLVALIIGFALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFL 417
IL+ G++ I F++G+G +PW+IMSEI P ++KG AGS+ W A V++ T NFL
Sbjct: 67 ILAFAGVLIYIAAFSIGLGSVPWVIMSEIFPIHLKGTAGSLVVLVAWLGAWVVSYTFNFL 246
Query: 418 LNWSSAGTFTIYAIFSAFTVAFALLFVPETKDRTLEEIQA 457
++WSS GT +YA S T+ F VPETK +TLEEIQA
Sbjct: 247 MSWSSPGTLFLYAGCSLLTILFVAKLVPETKGKTLEEIQA 366
>TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporter 4, partial
(54%)
Length = 1142
Score = 101 bits (251), Expect = 8e-22
Identities = 76/264 (28%), Positives = 129/264 (48%), Gaps = 12/264 (4%)
Frame = +3
Query: 204 ESPRWLAEMGLMERFESSLQTLRGSN---------VDITMEAQEIQNRFYTVCRPQEEKI 254
++P L E G +E ++ L+ +RG++ V+ + A+E+++ F + + +
Sbjct: 3 DTPNSLIERGRLEEGKTVLKKIRGTDNIELEFQELVEASRVAKEVKHPFRNLLKRRNRPQ 182
Query: 255 LVSFDGIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGL---GAIQVVMTGVATW 311
LV I L + QQ +GIN + FYA +F + G + DA+ + GA+ V+ T V+ +
Sbjct: 183 LVI--SIALQIFQQFTGINAIMFYAPVLFNTLGFKN-DASLYSAVITGAVNVLSTVVSIY 353
Query: 312 LVDKSGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGF 371
VDK GRR+LL+ + M +S +++A ++ V +L + IL VV + + F
Sbjct: 354 SVDKLGRRMLLLEAGVQMFLSQVVIAIILGIK-VTDHSDDLSKGIAILVVVMVCTFVSSF 530
Query: 372 ALGIGPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAI 431
A GP+ WLI SE P + SV N VI +L G F ++
Sbjct: 531 AWSWGPLGWLIPSETFPLETRSAGQSVTVCVNLLFTFVIAQAFLSMLCHFKFGIFLFFSG 710
Query: 432 FSAFTVAFALLFVPETKDRTLEEI 455
+ F L +PETK+ +EE+
Sbjct: 711 WVLVMSVFVLFLLPETKNVPIEEM 782
>CA952974 similar to GP|17381265|gb| AT5g18840/F17K4_90 {Arabidopsis
thaliana}, partial (19%)
Length = 372
Score = 97.8 bits (242), Expect(2) = 1e-21
Identities = 45/94 (47%), Positives = 64/94 (67%)
Frame = +3
Query: 69 FSLFGSLSNVGAMVGATVSGHLAEYFGRKGSLLVAAVPNIFGWVAISLAKDSSLLFIGRL 128
FS+FGSL +GAM+GA SG + ++ GRKG++ ++A I GW+ + +K S L +GR
Sbjct: 12 FSMFGSLVTIGAMLGAISSGRITDFIGRKGAMRISAGFCITGWLVVFFSKGSYSLDLGRF 191
Query: 129 LEGFGVGIISYVVPVYIAEIPPRTMRGSLGAVNQ 162
G+G+G+ISYVVPVYI EI P+ +R L NQ
Sbjct: 192 FTGYGIGVISYVVPVYIVEIAPKNLREELATTNQ 293
Score = 23.5 bits (49), Expect(2) = 1e-21
Identities = 9/17 (52%), Positives = 10/17 (57%)
Frame = +2
Query: 176 GLFFNWRTLALLGVFPC 192
G NWR LAL G+ C
Sbjct: 293 GSVINWRKLALAGLVSC 343
>TC216286 similar to UP|Q8GTR0 (Q8GTR0) Sugar transporter, partial (45%)
Length = 1342
Score = 100 bits (248), Expect = 2e-21
Identities = 73/209 (34%), Positives = 112/209 (52%), Gaps = 4/209 (1%)
Frame = +2
Query: 256 VSFDGIGLLVLQQLSGINGVFFYASKIFASAGISSSDAATFGLGAIQVVMTGVATWLVDK 315
V F G L LQQLSGIN VF+++S +F SAG+ SD A +G + + V+ L+DK
Sbjct: 116 VVFIGSTLFALQQLSGINAVFYFSSIVFKSAGV-PSDIANVCIGIANLAGSIVSMGLMDK 292
Query: 316 SGRRILLIVSSSIMTISLLLVASAFYLEGVVSKGSELHNILGILSVVGLVALIIGFALGI 375
GR++LL S M I+++L A+ G S S + SV G+ ++ FALG
Sbjct: 293 LGRKVLLFWSFFGMAIAMILQAT-----GATSLVSNMG--AQYFSVGGMFLFVLTFALGA 451
Query: 376 GPIPWLIMSEILPPNIKGLAGSVATFSNWFTASVITLTANFLLNWSSAGTFTIYAIFSAF 435
GP+P L++ EI P I+ A +V +W + L FL G +Y++F+ F
Sbjct: 452 GPVPGLLLPEIFPSRIRAKAMAVCMSVHWVINFFVGLL--FLRLLEKLGPQLLYSMFATF 625
Query: 436 TVAFALLFVP----ETKDRTLEEIQASFI 460
+ A++FV ETK ++L EI+ + +
Sbjct: 626 CI-MAVIFVKRNVVETKGKSLHEIEIALL 709
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.141 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,343,986
Number of Sequences: 63676
Number of extensions: 258885
Number of successful extensions: 1770
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 1682
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1702
length of query: 461
length of database: 12,639,632
effective HSP length: 101
effective length of query: 360
effective length of database: 6,208,356
effective search space: 2235008160
effective search space used: 2235008160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0283.14


