

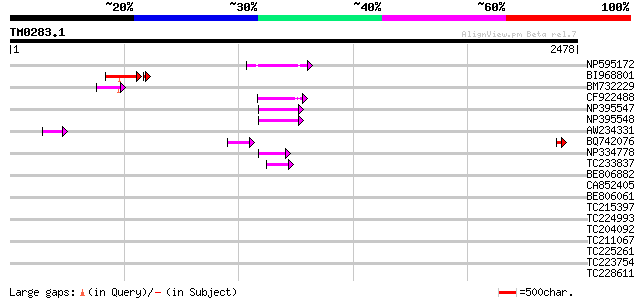
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0283.1
(2478 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 164 4e-40
BI968801 similar to GP|24461860|gb CTV.20 {Poncirus trifoliata},... 137 5e-37
BM732229 103 7e-22
CF922488 99 2e-20
NP395547 reverse transcriptase [Glycine max] 97 7e-20
NP395548 reverse transcriptase [Glycine max] 84 1e-15
AW234331 75 4e-13
BQ742076 70 1e-11
NP334778 reverse transcriptase [Glycine max] 70 1e-11
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 62 2e-09
BE806882 similar to GP|27764548|gb polyprotein {Glycine max}, pa... 42 0.003
CA852405 weakly similar to GP|24461860|gb CTV.20 {Poncirus trifo... 38 0.048
BE806061 similar to GP|21595125|gb endosperm-specific protein-li... 37 0.14
TC215397 weakly similar to UP|Q8S9J6 (Q8S9J6) AT5g10770/T30N20_4... 36 0.24
TC224993 homologue to UP|Q9W3A9 (Q9W3A9) CG12660-PA, partial (15%) 33 1.2
TC204092 homologue to UP|PSAD_NICSY (P29302) Photosystem I react... 33 1.5
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 33 1.5
TC225261 homologue to UP|RS14_LUPLU (O22584) 40S ribosomal prote... 33 1.5
TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequen... 33 1.5
TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxida... 33 1.5
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 164 bits (415), Expect = 4e-40
Identities = 98/297 (32%), Positives = 159/297 (52%), Gaps = 5/297 (1%)
Frame = +1
Query: 1033 PTKARPIQMNEQYLKYCREEIGEYLNKGLIRHSKSPWSCIGFYVLNASELERGALRLVIN 1092
P K RP + + I E L +G+I+ S SP+S V + + G+ R +
Sbjct: 1789 PVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSPFSLPILLV----KKKDGSWRFCTD 1956
Query: 1093 YKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAFVVPFG 1152
Y+ LN + +P+P +L+ LH FSK D++SGY+QI V+ EDR KTAF G
Sbjct: 1957 YRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHG 2136
Query: 1153 HYEWNVMPQGLKNAPSEYQNIMNDIF-YPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIE 1211
HYEW VMP GL NAP+ +Q +MN IF + F +V+ DD+L++S H++HLE ++
Sbjct: 2137 HYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQ 2316
Query: 1212 IIKKNGLVVSAKKMKIFETKTRFLGYEI----YQGQITPIQRSLEFAKKFPDELKEKTQL 1267
+K++ L K +T+ +LG+++ + T +Q L++ P+ +K QL
Sbjct: 2317 TLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPT--PNNVK---QL 2481
Query: 1268 QRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSPPWSEEMSHSVREVKKLVQSLPIVT 1324
+ FLG Y FI + + PL L+K+S W+ E + ++KK + P+++
Sbjct: 2482 RGFLGLTGYYRRFIKSYANIAGPLTDLLQKDSFLWNNEAEAAFVKLKKAMTEAPVLS 2652
Score = 32.0 bits (71), Expect = 3.4
Identities = 24/81 (29%), Positives = 38/81 (46%), Gaps = 15/81 (18%)
Frame = +1
Query: 650 PYEQNPSTQQPTNKKNVNPKK-----------KNVCWKCGKPGHYANKCKTQQ----KIN 694
P PST+ P N +N N KK KN+C+ C + A+KC +Q ++
Sbjct: 877 PLLPTPSTK-PFNLRNQNIKKISPAEIQLRREKNLCYFCDEKFSPAHKCPNRQVMLLQLE 1053
Query: 695 ELDLDQKLKDSLISVLINSED 715
E D DQ + +++ N +D
Sbjct: 1054ETDEDQTDEQVMVTEEANMDD 1116
>BI968801 similar to GP|24461860|gb CTV.20 {Poncirus trifoliata}, partial
(0%)
Length = 660
Score = 137 bits (346), Expect(2) = 5e-37
Identities = 74/170 (43%), Positives = 104/170 (60%), Gaps = 13/170 (7%)
Frame = -1
Query: 420 MIMAATAYKVHNNTDRNAAL-MITHGFTGQLRGWWDNIMTPEDKSTILEKKKE--NGEE- 475
M M ATAY+ + + ++ GF+GQL+ WWDN +T E+K I K NG+
Sbjct: 654 MTMVATAYQTSHECSXXXIIDILVAGFSGQLKRWWDNYLTNEEK*KIYSAVKTDLNGKVI 475
Query: 476 ---------DAVATLIYTIILHFIGDPSIFRERASSQLSNLYCPTMSDYRWYKDTFFSKV 526
DAV TLI+TI HFIGDPS++++R++ LSNL C T++D+RWY+DTF ++V
Sbjct: 474 TNDDDKEILDAVNTLIFTIAQHFIGDPSLWKDRSAELLSNLKCRTLADFRWYRDTFLTRV 295
Query: 527 TLREDGNNAFWKERFIAGLPRLMQSKVLDNLSLFNQGNPINFGSLSFGQL 576
RED F KE+F+AGLPR + KV D + + I + SLS+GQL
Sbjct: 294 YTREDSQQPF*KEKFLAGLPRSLGDKVRDKIRSQSANGDIPYESLSYGQL 145
Score = 37.7 bits (86), Expect(2) = 5e-37
Identities = 13/31 (41%), Positives = 24/31 (76%)
Frame = -2
Query: 586 QVCTDFKLQNKMQKDMQISRKEVGSFCEQYG 616
++C D K+Q ++ K+ ++K++GSFCEQ+G
Sbjct: 116 KICQDDKIQRQLAKEKAQTKKDLGSFCEQFG 24
>BM732229
Length = 421
Score = 103 bits (258), Expect = 7e-22
Identities = 59/139 (42%), Positives = 82/139 (58%), Gaps = 13/139 (9%)
Frame = -3
Query: 381 DMRFEERREFVQNTFSGTSLDEWNIDGFSEQNILDITHQMIMAATAYKV-HNNTDRNAAL 439
D+ EER E +FS ++ EWNID +E NI++ M M ATAY+ H ++
Sbjct: 419 DLLLEERGENNFKSFSANNIYEWNIDAQTEYNIMNTLQHMTMVATAYQTSHECSEETIID 240
Query: 440 MITHGFTGQLRGWWDNIMTPEDKSTILEKKKE--NGE----------EDAVATLIYTIIL 487
++ GF+GQL+GWWDN +T E+KS I K NG+ DAV TLI+TI
Sbjct: 239 ILVAGFSGQLKGWWDNYLTNEEKSKIYSAVKTDLNGKVITNDDDKEIPDAVNTLIFTIAQ 60
Query: 488 HFIGDPSIFRERASSQLSN 506
HFIGDPS++ +R++ LSN
Sbjct: 59 HFIGDPSLW*DRSAELLSN 3
>CF922488
Length = 741
Score = 99.4 bits (246), Expect = 2e-20
Identities = 74/225 (32%), Positives = 115/225 (50%), Gaps = 9/225 (4%)
Frame = +3
Query: 1083 ERGALRLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDR 1142
E G + + ++Y+ LN ++PLP+ L+ + FS D SGY QI + ED
Sbjct: 12 EDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAPEDM 191
Query: 1143 YKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTI-VYLDDVLVFSKGIDQ 1201
KT F+ +G + + M GLKN + YQ M +F M+ I VY+DD++V S+ ++
Sbjct: 192 EKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRTEEE 371
Query: 1202 HVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFL--------GYEIYQGQITPIQRSLEF 1253
H+ +L K ++K L ++ K +FE K+R L G E+ ++ I LE
Sbjct: 372 HLVNLRKLFRRLRKYRLRLNPAKC-MFEVKSRKLLDFIDS*RGIEVDSNKVKVI---LEM 539
Query: 1254 AKKFPDELKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKN 1298
AK ++ Q+Q FLG +NY+ FI + C PLF L KN
Sbjct: 540 AKPHTEK-----QVQGFLGRLNYIVRFIS*LIATCEPLFILLCKN 659
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 97.4 bits (241), Expect = 7e-20
Identities = 62/195 (31%), Positives = 101/195 (51%), Gaps = 1/195 (0%)
Frame = +1
Query: 1088 RLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAF 1147
R+ I+Y+ LN+ + YPLP ++KRL + + + D SGY QI+V +D+ KTAF
Sbjct: 163 RMCIDYRKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAF 342
Query: 1148 VVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTI-VYLDDVLVFSKGIDQHVQHL 1206
PF + + MP GL NA + +Q M IF + I V++DD F + +L
Sbjct: 343 TCPFSVFAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANL 522
Query: 1207 EKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQ 1266
EK ++ +K+ LV++ +K + LG++I + I ++ L+ K P + K
Sbjct: 523 EKVLQRCEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVK-G 699
Query: 1267 LQRFLGCVNYVADFI 1281
+ FLG V + FI
Sbjct: 700 IHSFLGHVGFYRRFI 744
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 83.6 bits (205), Expect = 1e-15
Identities = 57/195 (29%), Positives = 97/195 (49%), Gaps = 1/195 (0%)
Frame = +1
Query: 1088 RLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAF 1147
+L I+Y+ LN+ + +PLP +++RL + D GY QI V +D+ K AF
Sbjct: 163 KLCIDYRKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAF 342
Query: 1148 VVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTI-VYLDDVLVFSKGIDQHVQHL 1206
PFG + + +P GL NAP+ +Q M IF + +I V++DD VF ++ ++ L
Sbjct: 343 TCPFGVFAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKL 522
Query: 1207 EKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQGQITPIQRSLEFAKKFPDELKEKTQ 1266
E ++ + LV++ +K + LG++I I Q ++ +K P K
Sbjct: 523 EMVLQRCVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVK-G 699
Query: 1267 LQRFLGCVNYVADFI 1281
++ FLG + FI
Sbjct: 700 IRSFLGQARFYRRFI 744
>AW234331
Length = 370
Score = 75.1 bits (183), Expect = 4e-13
Identities = 36/112 (32%), Positives = 67/112 (59%), Gaps = 3/112 (2%)
Frame = +2
Query: 145 SFEINKDFIRKYFMAEYNKDKRLWYFKNFSKLETENLRTLYYEEMEVSETNIYFFD*FEN 204
+F I+K+ +RK F + N+ +R W+F+++ + ++ +Y+ +E + NI FFD F
Sbjct: 8 NFSIDKETLRKDFYSSENEPQRRWFFQHYKGTNRKQIKDKFYKFVERVKINILFFDWFHA 187
Query: 205 YCIKNNLNYPFWKNV--NPITKIDTIWKTPDNNTITSEYPPQTGVKIA-IKD 253
Y I+ +++YP+ +++ +P T + T WK D I SE PP T ++ +KD
Sbjct: 188 YAIRKDIDYPWKQDIIGDPTTNVITNWKIKDGELIQSELPPATQYQLPNVKD 343
>BQ742076
Length = 429
Score = 70.1 bits (170), Expect = 1e-11
Identities = 44/128 (34%), Positives = 71/128 (55%), Gaps = 12/128 (9%)
Frame = -2
Query: 953 QFYEKTKEGVNSANGSKMNIQYKLSNAKICKNQICFKSSFVLVKN--MTEKV------IL 1004
+ E K ++ ++ SK+ + K++ K+++ F + + ++ M EK+ I
Sbjct: 389 ELIET*KSNLHRSSTSKLPSKKKINQINFLKDEVSFSNIQIQLEKPQMKEKISSLLKHIQ 210
Query: 1005 GTLFICLLYP----KVHTIALPYIDEFNEKDIPTKARPIQMNEQYLKYCREEIGEYLNKG 1060
T+ L K H + LP +F EK IPTK+RPI MNE+ L+YC++EI + NKG
Sbjct: 209 STICFDLARAF*NRKKHILNLPCDKDFKEKHIPTKSRPI*MNEELLQYCQKEIKDLFNKG 30
Query: 1061 LIRHSKSP 1068
LIR SK+P
Sbjct: 29 LIRKSKNP 6
Score = 53.9 bits (128), Expect = 8e-07
Identities = 22/42 (52%), Positives = 35/42 (82%)
Frame = -2
Query: 2389 KEKQINFLKEEISYKNIEVQLQQPSVKSRIENILGNIQSSIC 2430
K QINFLK+E+S+ NI++QL++P +K +I ++L +IQS+IC
Sbjct: 323 KINQINFLKDEVSFSNIQIQLEKPQMKEKISSLLKHIQSTIC 198
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 69.7 bits (169), Expect = 1e-11
Identities = 41/138 (29%), Positives = 74/138 (52%), Gaps = 1/138 (0%)
Frame = +3
Query: 1088 RLVINYKPLNKVLKWIRYPLPNKTDLIKRLHKGTIFSKFDMKSGYYQISVREEDRYKTAF 1147
R+ ++Y+ LN+ +PLP+ L+ + +FS D SGY QI + ED KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 1148 VVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYMNFTI-VYLDDVLVFSKGIDQHVQHL 1206
+ +G + + VM GLKN + Y M +F M+ I Y+D+++ S+ ++H+ +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 1207 EKFIEIIKKNGLVVSAKK 1224
+ ++K L ++ +K
Sbjct: 363 QNLFGQLRKYRLRLNPRK 416
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 62.4 bits (150), Expect = 2e-09
Identities = 41/120 (34%), Positives = 63/120 (52%), Gaps = 1/120 (0%)
Frame = +2
Query: 1123 FSKFDMKSGYYQISVREEDRYKTAFVVPFGHYEWNVMPQGLKNAPSEYQNIMNDIFYPYM 1182
FS D SGY QI + ED KT FV +G + + VM GLKN + YQ M +F+ M
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 1183 NFTI-VYLDDVLVFSKGIDQHVQHLEKFIEIIKKNGLVVSAKKMKIFETKTRFLGYEIYQ 1241
+ I VY+DD++ S+ +H+ +L K ++K L ++ K + LG+ + Q
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
>BE806882 similar to GP|27764548|gb polyprotein {Glycine max}, partial (3%)
Length = 153
Score = 42.0 bits (97), Expect = 0.003
Identities = 19/51 (37%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Frame = -1
Query: 1163 LKNAPSEYQNIMNDIF-YPYMNFTIVYLDDVLVFSKGIDQHVQHLEKFIEI 1212
L NAP+ +Q +MN IF + + +V+ DD+LV+S +H+ HLE ++
Sbjct: 153 LTNAPTSFQCLMNHIFQHALRKYVLVFFDDILVYSSTWHEHLCHLEVVFKV 1
>CA852405 weakly similar to GP|24461860|gb CTV.20 {Poncirus trifoliata},
partial (0%)
Length = 674
Score = 38.1 bits (87), Expect = 0.048
Identities = 20/56 (35%), Positives = 33/56 (58%), Gaps = 2/56 (3%)
Frame = +3
Query: 1477 YEFILVDSDSIEVTH--NPDKQGEIAFSKIKIIKVLSAQDWNAPLHYFKNFSRQFD 1530
YEFIL+D++S+ + H +P S I+I+KV+ + + + L+ K FS FD
Sbjct: 30 YEFILIDTNSVSMKHFKDPKDPNLNTHSTIQILKVMQPRHYGSNLNQPKKFSAPFD 197
>BE806061 similar to GP|21595125|gb endosperm-specific protein-like protein
{Arabidopsis thaliana}, partial (26%)
Length = 355
Score = 36.6 bits (83), Expect = 0.14
Identities = 30/98 (30%), Positives = 43/98 (43%), Gaps = 11/98 (11%)
Frame = +3
Query: 1351 PPLGSSSTSKDKALAITPLKSAGSSAPATPSKSSQ-RLTTSQIVKTPP-----QNQKAPL 1404
PP + S + T +A S PA+P+KS+ R + S TPP QN +P
Sbjct: 27 PPPPTISPTFSPPTPTTATSTASSPKPASPAKSTPARPSPSSSSTTPPSPPWPQNTPSPS 206
Query: 1405 VPLAN-----KYTVLTQSSSKSPVKPESTEYFEKPEGL 1437
++ T TQSS++SP P S KP +
Sbjct: 207 AKTSSASTSSSTTSTTQSSTRSPTAPRSPPLSTKPPAM 320
>TC215397 weakly similar to UP|Q8S9J6 (Q8S9J6) AT5g10770/T30N20_40, partial
(76%)
Length = 1447
Score = 35.8 bits (81), Expect = 0.24
Identities = 30/90 (33%), Positives = 37/90 (40%), Gaps = 11/90 (12%)
Frame = +1
Query: 1350 *PPLGSSSTSKDKALAITPLKSAGSSAP----ATPSKSSQRLTTSQIVKTPPQNQKAPLV 1405
*PP SS+TS A T SA A ATPS SS +L + + +P + P
Sbjct: 421 *PPPTSSTTSSSAAARTTRASSAAPPASLASAATPSPSSNKLPPNTVKSSPTASPPPPAP 600
Query: 1406 PLAN-------KYTVLTQSSSKSPVKPEST 1428
P + T T S SP P ST
Sbjct: 601 PATSPSAPPPQAATSSTPPSPPSPAAPPST 690
>TC224993 homologue to UP|Q9W3A9 (Q9W3A9) CG12660-PA, partial (15%)
Length = 526
Score = 33.5 bits (75), Expect = 1.2
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +1
Query: 674 CWKCGKPGHYANKCKTQQKI 693
C+ CGKPGH A+ C TQ+++
Sbjct: 91 CFNCGKPGHRASDCITQKRV 150
>TC204092 homologue to UP|PSAD_NICSY (P29302) Photosystem I reaction center
subunit II, chloroplast precursor (Photosystem I 20 kDa
subunit) (PSI-D), partial (85%)
Length = 934
Score = 33.1 bits (74), Expect = 1.5
Identities = 18/68 (26%), Positives = 34/68 (49%)
Frame = +2
Query: 1980 YANNLIFKVVPRNLNPESKILLNNNGKGGLAGMIIGNPNLDQNPIPRKLQKTLLRIDPLK 2039
+ NL+ + + NP + I+ +N+G G NP+L NP P + Q R+ ++
Sbjct: 56 HIQNLLLYSLSLSDNPNTCIVSSNHGNG--------NPSLSLNPTPLRSQSQRPRLRAME 211
Query: 2040 EKMSPAII 2047
K+ P ++
Sbjct: 212 AKLQPLLL 235
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 33.1 bits (74), Expect = 1.5
Identities = 16/64 (25%), Positives = 32/64 (50%), Gaps = 1/64 (1%)
Frame = +1
Query: 1261 LKEKTQLQRFLGCVNYVADFIPNIRIVCAPLFKRLRKNSP-PWSEEMSHSVREVKKLVQS 1319
LK ++ F G ++ F+PN V +PL + ++KN W E+ + +K+ +
Sbjct: 103 LKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTK 282
Query: 1320 LPIV 1323
P++
Sbjct: 283 APVL 294
>TC225261 homologue to UP|RS14_LUPLU (O22584) 40S ribosomal protein S14,
complete
Length = 849
Score = 33.1 bits (74), Expect = 1.5
Identities = 19/32 (59%), Positives = 21/32 (65%)
Frame = -2
Query: 1834 LMCINRHHRAEEKASGFHLPFSLSLSLSLSLS 1865
L C R R +E +S F L SLSLSLSLSLS
Sbjct: 146 LQCD*RIRRLDEPSSAFTLSLSLSLSLSLSLS 51
>TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequence, partial
(22%)
Length = 742
Score = 33.1 bits (74), Expect = 1.5
Identities = 10/17 (58%), Positives = 14/17 (81%)
Frame = +2
Query: 671 KNVCWKCGKPGHYANKC 687
K+ C+ CGKPGH+A +C
Sbjct: 413 KSTCFNCGKPGHFAREC 463
>TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxidase ATP26a
{Arabidopsis thaliana;} , partial (49%)
Length = 731
Score = 33.1 bits (74), Expect = 1.5
Identities = 23/78 (29%), Positives = 36/78 (45%)
Frame = +3
Query: 1351 PPLGSSSTSKDKALAITPLKSAGSSAPATPSKSSQRLTTSQIVKTPPQNQKAPLVPLANK 1410
P SSST+ A+TP S S +P S+ +TS TP + AP PL++
Sbjct: 189 PRSASSSTTASSPTAVTPPSS--SPPRPSPGPSATPTSTSPSPATPSTSSSAPRPPLSSP 362
Query: 1411 YTVLTQSSSKSPVKPEST 1428
+ + + SP P ++
Sbjct: 363 APTPSPAPTFSPPPPATS 416
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 108,523,990
Number of Sequences: 63676
Number of extensions: 1619356
Number of successful extensions: 17622
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 10175
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 15605
length of query: 2478
length of database: 12,639,632
effective HSP length: 113
effective length of query: 2365
effective length of database: 5,444,244
effective search space: 12875637060
effective search space used: 12875637060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0283.1


