

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
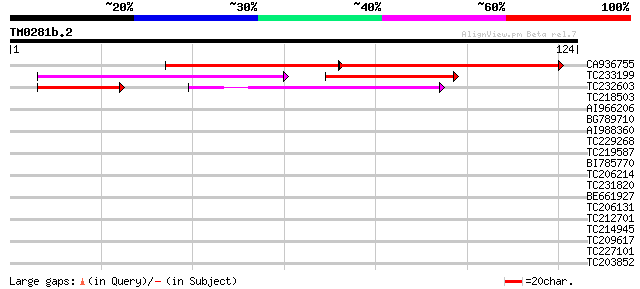
Query= TM0281b.2
(124 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CA936755 60 2e-17
TC233199 52 1e-11
TC232603 weakly similar to UP|Q39482 (Q39482) Legumin (Fragment)... 40 3e-07
TC218503 28 1.4
AI966206 27 3.2
BG789710 homologue to GP|532754|gb|AA RNase NE {Nicotiana alata}... 26 4.1
AI988360 26 4.1
TC229268 similar to GB|BAB32889.1|13122296|AB047812 hin1 homolog... 26 5.4
TC219587 similar to PIR|T49181|T49181 cyclophylin-like protein -... 26 5.4
BI785770 similar to PIR|A96842|A968 F23A5.27 [imported] - Arabid... 26 5.4
TC206214 beta-D-glucan exohydrolase [Glycine max] 26 5.4
TC231820 similar to UP|Q9SAG9 (Q9SAG9) F23A5.27, partial (26%) 26 5.4
BE661927 26 5.4
TC206131 similar to UP|Q9LYF9 (Q9LYF9) Clathrin binding protein-... 26 5.4
TC212701 similar to UP|Q76KV2 (Q76KV2) DNA binding with one fing... 25 7.0
TC214945 UP|Q819B7 (Q819B7) Stage V sporulation protein AB, part... 25 7.0
TC209617 25 7.0
TC227101 homologue to UP|WR23_ARATH (O22900) Probable WRKY trans... 25 9.2
TC203852 homologue to UP|PSBO_PEA (P14226) Oxygen-evolving enhan... 25 9.2
>CA936755
Length = 424
Score = 60.5 bits (145), Expect(2) = 2e-17
Identities = 21/49 (42%), Positives = 32/49 (64%)
Frame = -2
Query: 73 FLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPRPFRVLNCW 121
FL+S W WP Q + R SDHCP++ ++ ++WGP+PFRV++ W
Sbjct: 294 FLVSEQWLLSWPDSSQYTLPRDFSDHCPIIMQTKKVDWGPKPFRVVDWW 148
Score = 43.5 bits (101), Expect(2) = 2e-17
Identities = 18/39 (46%), Positives = 25/39 (63%)
Frame = -1
Query: 35 MADFKAFLSNLSLIEPPAVGKKYTWFRPNGQAASRLDRF 73
+++F ++S + L E VG YTW RPN + SRLDRF
Sbjct: 409 ISEFNTWISEMELQEIKCVGSTYTWIRPNARVKSRLDRF 293
>TC233199
Length = 1105
Score = 51.6 bits (122), Expect(2) = 1e-11
Identities = 23/55 (41%), Positives = 33/55 (59%)
Frame = +2
Query: 7 FCIAGDFNVVARSEERMGISDSIYGRSDMADFKAFLSNLSLIEPPAVGKKYTWFR 61
+CI DFN V R ER+G S G M +F ++ +L + + P+VG KYTW+R
Sbjct: 290 WCIVDDFNCVRRQSERVGASQREQGGGIMTEFNDWIVDLEVEDMPSVGHKYTWYR 454
Score = 32.7 bits (73), Expect(2) = 1e-11
Identities = 13/29 (44%), Positives = 19/29 (64%)
Frame = +1
Query: 70 LDRFLLSSDWCSCWPHGVQTVMARSLSDH 98
LDR L+S +W S W +Q ++ R+ SDH
Sbjct: 466 LDRVLVSVEWFSKWSGSIQFILDRNYSDH 552
>TC232603 weakly similar to UP|Q39482 (Q39482) Legumin (Fragment), partial
(4%)
Length = 1123
Score = 40.0 bits (92), Expect(2) = 3e-07
Identities = 19/56 (33%), Positives = 33/56 (58%)
Frame = +2
Query: 40 AFLSNLSLIEPPAVGKKYTWFRPNGQAASRLDRFLLSSDWCSCWPHGVQTVMARSL 95
+F+ NL+L VG+KY W++ + SRLDRFL+ + + WP+ ++ R +
Sbjct: 518 SFVVNLAL-----VGRKYNWYKDDKTTMSRLDRFLVYEE*LTTWPNMTLILLLRRI 670
Score = 29.3 bits (64), Expect(2) = 3e-07
Identities = 11/19 (57%), Positives = 13/19 (67%)
Frame = +1
Query: 7 FCIAGDFNVVARSEERMGI 25
+CI GDFN V EER G+
Sbjct: 430 WCIVGDFNAVCSEEERKGV 486
>TC218503
Length = 640
Score = 27.7 bits (60), Expect = 1.4
Identities = 13/30 (43%), Positives = 16/30 (53%)
Frame = +2
Query: 71 DRFLLSSDWCSCWPHGVQTVMARSLSDHCP 100
DR LS D S + QT+ +SLS CP
Sbjct: 182 DREFLSQDIASAFQEQTQTLQQQSLSSECP 271
>AI966206
Length = 502
Score = 26.6 bits (57), Expect = 3.2
Identities = 12/29 (41%), Positives = 16/29 (54%)
Frame = -3
Query: 7 FCIAGDFNVVARSEERMGISDSIYGRSDM 35
+CI DFN V ++ ER+G S G M
Sbjct: 89 WCIIEDFNCVCKNSERVGASQRDQGSGVM 3
>BG789710 homologue to GP|532754|gb|AA RNase NE {Nicotiana alata}, partial
(9%)
Length = 407
Score = 26.2 bits (56), Expect = 4.1
Identities = 12/35 (34%), Positives = 19/35 (54%)
Frame = +2
Query: 49 EPPAVGKKYTWFRPNGQAASRLDRFLLSSDWCSCW 83
E +G+++T + A RL LLS DW +C+
Sbjct: 86 ERKTIGQEHTVTQSRAAAILRLGNLLLS*DWETCY 190
>AI988360
Length = 401
Score = 26.2 bits (56), Expect = 4.1
Identities = 8/14 (57%), Positives = 12/14 (85%)
Frame = -1
Query: 53 VGKKYTWFRPNGQA 66
+G+K+TW+R NG A
Sbjct: 221 IGRKFTWYRSNGLA 180
>TC229268 similar to GB|BAB32889.1|13122296|AB047812 hin1 homolog
{Arabidopsis thaliana;} , partial (31%)
Length = 1043
Score = 25.8 bits (55), Expect = 5.4
Identities = 17/77 (22%), Positives = 32/77 (41%), Gaps = 2/77 (2%)
Frame = -3
Query: 12 DFNVVARSEERMGISDSIYGRSDM--ADFKAFLSNLSLIEPPAVGKKYTWFRPNGQAASR 69
DF+ + +G + ++ ++ K+F S + EP ++ TWF +G
Sbjct: 735 DFSGTCNAHRILGFTLALLNSKNLNRTTIKSFKSMV*TPEPFSLSNTSTWFPASGVTRWP 556
Query: 70 LDRFLLSSDWCSCWPHG 86
L R + CW +G
Sbjct: 555 LKRGFTKVVFLWCWKNG 505
>TC219587 similar to PIR|T49181|T49181 cyclophylin-like protein - Arabidopsis
thaliana {Arabidopsis thaliana;} , partial (19%)
Length = 1344
Score = 25.8 bits (55), Expect = 5.4
Identities = 13/52 (25%), Positives = 23/52 (44%)
Frame = -1
Query: 59 WFRPNGQAASRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNW 110
W + AA F+++ WCS W G + +A P +++S +W
Sbjct: 1068 WRARSRPAARHGSPFIITFWWCSIWGSGARRAIAPRFGL*PPPVWQSFITSW 913
>BI785770 similar to PIR|A96842|A968 F23A5.27 [imported] - Arabidopsis
thaliana, partial (15%)
Length = 433
Score = 25.8 bits (55), Expect = 5.4
Identities = 11/21 (52%), Positives = 14/21 (66%)
Frame = -1
Query: 64 GQAASRLDRFLLSSDWCSCWP 84
G+ A + R +LSS W SCWP
Sbjct: 133 GKPAGKKKR-ILSSSWPSCWP 74
>TC206214 beta-D-glucan exohydrolase [Glycine max]
Length = 1647
Score = 25.8 bits (55), Expect = 5.4
Identities = 7/11 (63%), Positives = 8/11 (72%)
Frame = +3
Query: 73 FLLSSDWCSCW 83
F+L S WC CW
Sbjct: 579 FILCSSWCQCW 611
>TC231820 similar to UP|Q9SAG9 (Q9SAG9) F23A5.27, partial (26%)
Length = 498
Score = 25.8 bits (55), Expect = 5.4
Identities = 11/21 (52%), Positives = 14/21 (66%)
Frame = -1
Query: 64 GQAASRLDRFLLSSDWCSCWP 84
G+ A + R +LSS W SCWP
Sbjct: 153 GKPAGKKKR-ILSSSWPSCWP 94
>BE661927
Length = 768
Score = 25.8 bits (55), Expect = 5.4
Identities = 13/35 (37%), Positives = 16/35 (45%)
Frame = -1
Query: 51 PAVGKKYTWFRPNGQAASRLDRFLLSSDWCSCWPH 85
P V +T F PNG + + LL CS W H
Sbjct: 660 PIVAATHTHFLPNGVSQKL*NYPLLHPQRCSXWEH 556
>TC206131 similar to UP|Q9LYF9 (Q9LYF9) Clathrin binding protein-like,
partial (52%)
Length = 2220
Score = 25.8 bits (55), Expect = 5.4
Identities = 14/52 (26%), Positives = 28/52 (52%), Gaps = 4/52 (7%)
Frame = +1
Query: 15 VVARSEERMGISDSIYGRS----DMADFKAFLSNLSLIEPPAVGKKYTWFRP 62
++ +S+++ G + G+ ++ D K+FL + IE PA+ T F+P
Sbjct: 37 LIPKSKKKQG*GKFLEGKQKFDPNVLDLKSFLLKTTAIEFPAINISITPFQP 192
>TC212701 similar to UP|Q76KV2 (Q76KV2) DNA binding with one finger 2
protein, partial (8%)
Length = 959
Score = 25.4 bits (54), Expect = 7.0
Identities = 11/34 (32%), Positives = 17/34 (49%)
Frame = -1
Query: 50 PPAVGKKYTWFRPNGQAASRLDRFLLSSDWCSCW 83
PP+ K+ W+ + + DR L + WC CW
Sbjct: 164 PPSQQKRSHWWLKRSRHSRFGDRSL-PATWCCCW 66
>TC214945 UP|Q819B7 (Q819B7) Stage V sporulation protein AB, partial (7%)
Length = 743
Score = 25.4 bits (54), Expect = 7.0
Identities = 11/34 (32%), Positives = 15/34 (43%)
Frame = +1
Query: 50 PPAVGKKYTWFRPNGQAASRLDRFLLSSDWCSCW 83
PP YT R N + A++ + S W CW
Sbjct: 286 PPTTS*TYTTTRDNNKGATKQRKERPSPSWT*CW 387
>TC209617
Length = 1092
Score = 25.4 bits (54), Expect = 7.0
Identities = 12/32 (37%), Positives = 16/32 (49%), Gaps = 2/32 (6%)
Frame = -1
Query: 83 WPHGVQTVMARSLSDHCPVM--FRSTDMNWGP 112
W H +Q M L HCPV+ + S + W P
Sbjct: 666 WHHQLQCEMVLQL*GHCPVLPFYSSKWLQWLP 571
>TC227101 homologue to UP|WR23_ARATH (O22900) Probable WRKY transcription
factor 23 (WRKY DNA-binding protein 23), partial (31%)
Length = 599
Score = 25.0 bits (53), Expect = 9.2
Identities = 11/24 (45%), Positives = 11/24 (45%)
Frame = -1
Query: 80 CSCWPHGVQTVMARSLSDHCPVMF 103
C CWP V T M SL D F
Sbjct: 284 CVCWPS*VVTTMLGSLKDRSTRFF 213
>TC203852 homologue to UP|PSBO_PEA (P14226) Oxygen-evolving enhancer protein
1, chloroplast precursor (OEE1) (33 kDa subunit of
oxygen evolving system of photosystem II) (OEC 33 kDa
subunit) (33 kDa thylakoid membrane protein), partial
(97%)
Length = 1606
Score = 25.0 bits (53), Expect = 9.2
Identities = 16/53 (30%), Positives = 22/53 (41%)
Frame = +3
Query: 61 RPNGQAASRLDRFLLSSDWCSCWPHGVQTVMARSLSDHCPVMFRSTDMNWGPR 113
RP+ + + F SS S W G QT + S H V ++W PR
Sbjct: 888 RPSQSSYREANVFRSSSPSSSWWHQGSQTALVESFWSHHTV----EALSWTPR 1034
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.137 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,305,854
Number of Sequences: 63676
Number of extensions: 99532
Number of successful extensions: 799
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 796
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 799
length of query: 124
length of database: 12,639,632
effective HSP length: 85
effective length of query: 39
effective length of database: 7,227,172
effective search space: 281859708
effective search space used: 281859708
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0281b.2


