

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0281a.3
(1430 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
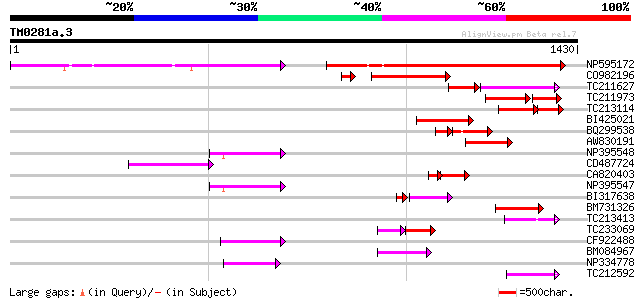
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 521 e-148
CO982196 255 2e-75
TC211627 130 4e-52
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 130 2e-48
TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, parti... 127 1e-41
BI425021 164 3e-40
BQ299538 114 6e-38
AW830191 144 4e-34
NP395548 reverse transcriptase [Glycine max] 122 1e-27
CD487724 121 2e-27
CA820403 weakly similar to GP|13273463|gb| pol protein integrase... 94 1e-25
NP395547 reverse transcriptase [Glycine max] 109 1e-23
BI317638 weakly similar to GP|9294238|dbj| contains similarity t... 95 3e-23
BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza... 107 4e-23
TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 98 2e-20
TC233069 78 6e-20
CF922488 96 1e-19
BM084967 95 2e-19
NP334778 reverse transcriptase [Glycine max] 92 2e-18
TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 91 5e-18
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 521 bits (1343), Expect = e-148
Identities = 274/606 (45%), Positives = 383/606 (62%), Gaps = 4/606 (0%)
Frame = +1
Query: 799 LDARGQTLAFWSQGLSPRAQHKSVYERELMALVQAIQKWRHYLMGRHFIVRTDQKSLKFL 858
L G +A++S+ L+PR Q +S Y REL+A+ +A+ K+RHYL+G FI+RTDQ+SLK L
Sbjct: 2719 LGQNGHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNKFIIRTDQRSLKSL 2898
Query: 859 TDQRLFS*DQFKWAVKLVGLDFEIQYKPGSENTVADALSRKMMYSALSILTSAEKEDWNS 918
DQ L + +Q W K +G DF+I+YKPG +N ADALSR M A S S E+ +
Sbjct: 2899 MDQSLQTPEQQAWLHKFLGYDFKIEYKPGKDNQAADALSRMFML-AWSEPHSIFLEELRA 3075
Query: 919 ELQQDSKLVKVIQDLLI-SPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISECHDTPT 977
L D L ++++ + SH + ++GLLY+K+R+V+ + I+ E H +P
Sbjct: 3076 RLISDPHLKQLMETYKQGADASH--YTVREGLLYWKDRVVIPAEEEIVNKILQEYHSSPI 3249
Query: 978 GGHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVW 1037
GGH+G RT R+ + YW M+ D+K +++ C ICQ++K + P GLLQPLPIP QVW
Sbjct: 3250 GGHAGITRTLARLKAQFYWPKMQEDVKAYIQKCLICQQAKSNNTLPAGLLQPLPIPQQVW 3429
Query: 1038 MDISMDFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELVKLHGFP 1097
D++MDF+ GLP G ++VV+DRLTK+AHF L Y++K VA F+ +VKLHG P
Sbjct: 3430 EDVAMDFITGLPNSFGLSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSHIVKLHGIP 3609
Query: 1098 TTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKP 1157
+IVSDRD +F S FW+ LFK+ GT L +S++YHPQ+DGQ+EV N+CLE YLRCF P
Sbjct: 3610 RSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYLRCFTYEHP 3789
Query: 1158 KQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRDG 1217
K W+ LPWAEFW+NT Y+ S GM+PF+ALYGR+PP L + EV + RD
Sbjct: 3790 KGWVKALPWAEFWYNTAYHMSLGMTPFRALYGREPPTLTRQACSIDDPAEVREQLTDRDA 3969
Query: 1218 VLGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFY 1277
+L +LK NL +AQ MK QADK R V F GD V +KLQPY+ S R KL+ R++
Sbjct: 3970 LLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQPYRQHSAVLRKNQKLSMRYF 4149
Query: 1278 GPYKILDRVGPVAYKLELPAHSKIHPVFHVSLLKQALKPTQQPQPLPPMLNEELELEVVP 1337
GP+K+L ++G VAYKLELP+ ++IHPVFHVS LK Q P P+ E+ + P
Sbjct: 4150 GPFKVLAKIGDVAYKLELPSAARIHPVFHVSQLKPFNGTAQDPYLPLPLTVTEMGPVMQP 4329
Query: 1338 EDVIDHREDAQGN---LEVLIKWEQLPTCDNTWELAAVIQDKFPLFPLEDKVKLLGGVLI 1394
++ R +G+ ++L++WE + TWE I+ +P F LEDKV G +
Sbjct: 4330 VKILASRIIIRGHNQIEQILVQWENGLQDEATWEDIEDIKASYPTFNLEDKVVFKGEGNV 4509
Query: 1395 GLGLSQ 1400
G+S+
Sbjct: 4510 TNGMSR 4527
Score = 467 bits (1201), Expect = e-131
Identities = 267/716 (37%), Positives = 399/716 (55%), Gaps = 21/716 (2%)
Frame = +1
Query: 2 KLEIPIFSGDDAVGWTQKLERYFAMREVTEDERMQATMVALEGRALSWFQWWERCNPSPT 61
KL+ P F G + + W K E++F + +R+ V L+ + W+Q ++ P +
Sbjct: 295 KLDFPRFDGKNVMDWIFKAEQFFDYYATPDADRLIIASVHLDQDVVPWYQMLQKTEPFSS 474
Query: 62 WEAFKLAVVRRFQPSMIQNPFELLLSLKQVGTVDEYVEEFEKYAGALKEIDHDFVRGIFL 121
W+AF A+ F PS P L L Q TV+EY +F + + + + F+
Sbjct: 475 WQAFTRALELDFGPSAYDCPRATLFKLNQSATVNEYYMQFTALVNRVDGLSAEAILDCFV 654
Query: 122 NGLKEEIKAEVRLIQK---------SLLIEQKNQIVTKKGVGFTRPSSAFRNNNYSKVVT 172
+GL+EEI +V+ ++ + L E+K K T+ S N S
Sbjct: 655 SGLQEEISRDVKAMEPRTLTKAVALAKLFEEKYTSPPK-----TKTFSNLARNFTSNTSA 819
Query: 173 VEPQNNTDQKQDSSSVGSVAMPSRNNAGIDSSRNRGGEFKHLTGAQMREKRDKNLCFRCD 232
+ T+QK D+ + + + RN+ K ++ A+++ +R+KNLC+ CD
Sbjct: 820 TQKYPPTNQKNDNPKPNLPPLLPTPSTKPFNLRNQN--IKKISPAEIQLRREKNLCYFCD 993
Query: 233 EPYSREHKCRNKQLRMIIMEDDDDADCGE---FEDALSGEFNSLQLSLCSMAGLTSTKSW 289
E +S HKC N+Q+ ++ +E+ D+ E + + + ++ LSL +M G +
Sbjct: 994 EKFSPAHKCPNRQVMLLQLEETDEDQTDEQVMVTEEANMDDDTHHLSLNAMRGSNGVGTI 1173
Query: 290 KMVGQIGNQSIVVLIDCGASHNFISHKLVEQLNLTVAETQTYTVEVGDGYKIRCKGKCSQ 349
+ GQ+G ++ +L+D G+S NFI ++ + L L V V VG+G + +G Q
Sbjct: 1174 RFTGQVGGIAVKILVDGGSSDNFIQPRVAQVLKLPVEPAPNLRVLVGNGQILSAEGIVQQ 1353
Query: 350 LGIMIQALEIVQNFYLFGLNGMDVVLGIEWLASLGDIKANFGKMELTLKRGKEVYTLKGD 409
L + IQ E+ YL ++G DV+LG WLA+LG A++ + L + + TL+G+
Sbjct: 1354 LPLHIQGQEVKVPVYLLQISGADVILGSTWLATLGPHVADYAALTLKFFQNDKFITLQGE 1533
Query: 410 PSLTKSELSYGALMQILLEEGEGLLVHCDTGEKVNKK--ECEIPEIITGV-------LLQ 460
+ SE + L + + C + + K+ E + ++ T + L
Sbjct: 1534 GN---SEATQAQLHHFRRLQNTKSIEECFAIQLIQKEVPEDTLKDLPTNIDPELAILLHT 1704
Query: 461 FDTVFQDPQGLPPRRRHDHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLEAGVIRP 520
+ VF P LPP+R DHAI LK+G+ +RPY+ PH QK+++EK++ EML G+I+P
Sbjct: 1705 YAQVFAVPASLPPQREQDHAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQP 1884
Query: 521 SISPFSSPIILVKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDL 580
S SPFS PI+LVKKKDG WR C DYRALN +T+ + FP+P ++ELLDEL GA+ FSKLDL
Sbjct: 1885 SNSPFSLPILLVKKKDGSWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDL 2064
Query: 581 KSGYHQIRMKDADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALV 640
+SGYHQI ++ D EKTAFRTH GHYE+LVMPFGLTNAP+TFQ LMN++ + LRKF LV
Sbjct: 2065 RSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLV 2244
Query: 641 FFDDILVYSPDLLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVSGEG 696
FFDDIL+YS H HL+ VL L H L A KCSFG T ++YLGH VSG G
Sbjct: 2245 FFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLG 2412
>CO982196
Length = 812
Score = 255 bits (651), Expect(2) = 2e-75
Identities = 118/201 (58%), Positives = 155/201 (76%)
Frame = +1
Query: 912 EKEDWNSELQQDSKLVKVIQDLLISPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISE 971
E DW E+Q +L ++ Q +L T PG+ + G LY+K+RLVLSK S +IP+++ E
Sbjct: 208 ELADWEEEIQAYLELYEIYQGILTKTTKKPGYAIRGGKLYFKDRLVLSKNSTKIPLLLKE 387
Query: 972 CHDTPTGGHSGHFRTYKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLP 1031
D+P GGHSG FRT+KR+A+ ++W+GMK +++V CEIC+R+K STL+P GLL LP
Sbjct: 388 LQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYVAACEICRRNKTSTLSPAGLL*LLP 567
Query: 1032 IPTQVWMDISMDFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKELV 1091
IPT+VW DISMDF+GGLPK +GKD +LVVVDRLTK+AHF+AL HPY+AK+VA LFIKELV
Sbjct: 568 IPTKVWTDISMDFIGGLPKAQGKDNILVVVDRLTKYAHFFALSHPYTAKEVAELFIKELV 747
Query: 1092 KLHGFPTTIVSDRDPLFLSNF 1112
+LHGFP +IVSD LF+S F
Sbjct: 748 RLHGFPASIVSDXXRLFMSLF 810
Score = 48.1 bits (113), Expect(2) = 2e-75
Identities = 21/35 (60%), Positives = 28/35 (80%)
Frame = +2
Query: 838 RHYLMGRHFIVRTDQKSLKFLTDQRLFS*DQFKWA 872
RHY +G+ FI+RT+ +S KFL +QRL S +QFKWA
Sbjct: 53 RHYPVGKKFIIRTN*RSSKFLNEQRLMSEEQFKWA 157
>TC211627
Length = 1034
Score = 130 bits (326), Expect(2) = 4e-52
Identities = 86/204 (42%), Positives = 111/204 (54%), Gaps = 4/204 (1%)
Frame = +3
Query: 1187 LYGRDPPLLLKAGAIPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEF 1246
+YG+ PP L A S VE V+ + + L L K Q MK AD HRR + F
Sbjct: 243 MYGKPPPALPLYSAGTSTVEAVDAILHSLATIHHTLTCRLQKYQDSMKRIADSHRRDLTF 422
Query: 1247 NEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELPAHSKIHPVFH 1306
N GDWVY++L PY+ SI + KL+ RFYGPY+I RVG VAY+L+LP SKIHP+FH
Sbjct: 423 NIGDWVYVRL*PYRQTSIQST-YTKLSKRFYGPYQIQARVGQVAYRLQLPPTSKIHPIFH 599
Query: 1307 VSLLKQALKP-TQQPQPLPPMLNEELELEVVPEDVIDHREDAQGN---LEVLIKWEQLPT 1362
VSLLK P + LPP L V P +D + D +VL++W L
Sbjct: 600 VSLLKVHHGPIPPELLALPPFSTTNHPL-VQPLQFLDWKMDESTTPPIPQVLVQWTNLAP 776
Query: 1363 CDNTWELAAVIQDKFPLFPLEDKV 1386
D TWE ++D ++ LEDKV
Sbjct: 777 EDTTWESWTQLKD---IYDLEDKV 839
Score = 95.1 bits (235), Expect(2) = 4e-52
Identities = 43/79 (54%), Positives = 56/79 (70%)
Frame = +2
Query: 1107 LFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPW 1166
+F+S W ELF ++GT+L+ ST+YHPQTDGQTEV NR LE YLR FV P+ W +L
Sbjct: 2 IFISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSL 181
Query: 1167 AEFWFNTNYNSSAGMSPFK 1185
AE +NT+ +S G SPF+
Sbjct: 182 AE*CYNTSVHSGIGFSPFE 238
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 130 bits (328), Expect(2) = 2e-48
Identities = 63/113 (55%), Positives = 87/113 (76%)
Frame = +1
Query: 1201 IPSRVEEVNQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYK 1260
IPSR+EEVN+L RDG+L L++NLLK+ M +K RR +E+ GD V+LK+QPY+
Sbjct: 1 IPSRLEEVNKLIIARDGLLATLRENLLKS*DIM*ANTNKRRRDIEYVVGD*VFLKMQPYR 180
Query: 1261 LKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELPAHSKIHPVFHVSLLKQA 1313
+S+A R KL+PRFY P+++ ++VG +AYKL+LP+H KIHPVFHVSLLK+A
Sbjct: 181 RRSLAKRINEKLSPRFYAPFQVFNKVGTIAYKLDLPSHIKIHPVFHVSLLKKA 339
Score = 82.4 bits (202), Expect(2) = 2e-48
Identities = 37/73 (50%), Positives = 53/73 (71%)
Frame = +3
Query: 1319 QPQPLPPMLNEELELEVVPEDVIDHREDAQGNLEVLIKWEQLPTCDNTWELAAVIQDKFP 1378
Q QPLPPML+E+ +L+ + V+D RE GN++VLI+W+ LP +N+WE A +Q+ F
Sbjct: 339 QSQPLPPMLSEDWKLQTYSDSVLDSRELQPGNVKVLIQWKNLPPSENSWESVAKLQEIFS 518
Query: 1379 LFPLEDKVKLLGG 1391
++ LEDKV LLGG
Sbjct: 519 IYHLEDKVSLLGG 557
>TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, partial (7%)
Length = 810
Score = 127 bits (320), Expect(2) = 1e-41
Identities = 61/102 (59%), Positives = 77/102 (74%)
Frame = +3
Query: 1233 MKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYK 1292
MK AD+ RR V + GDWVYLKLQPY+LKS+A + KL+PRFYGPY+I ++G VA++
Sbjct: 3 MKAYADRSRRAVTLSVGDWVYLKLQPYRLKSLAKKRNEKLSPRFYGPYQIKKQIGLVAFE 182
Query: 1293 LELPAHSKIHPVFHVSLLKQALKPTQQPQPLPPMLNEELELE 1334
L+LP KIHPVFH SLLK+A+ T PQPLP ML+E+L E
Sbjct: 183 LDLPPARKIHPVFHASLLKKAVAATANPQPLPLMLSEDLSSE 308
Score = 62.4 bits (150), Expect(2) = 1e-41
Identities = 33/65 (50%), Positives = 42/65 (63%)
Frame = +2
Query: 1332 ELEVVPEDVIDHREDAQGNLEVLIKWEQLPTCDNTWELAAVIQDKFPLFPLEDKVKLLGG 1391
EL V P +V ++ G EVLI+ E LP + TWE VI+++FP F LEDKV LLGG
Sbjct: 299 ELRVFPAEVKAVHNNSNGIAEVLIQLEDLPDFEATWESVEVIKEQFPSFHLEDKVTLLGG 478
Query: 1392 VLIGL 1396
VL+ L
Sbjct: 479 VLLDL 493
>BI425021
Length = 426
Score = 164 bits (414), Expect = 3e-40
Identities = 75/142 (52%), Positives = 104/142 (72%)
Frame = -1
Query: 1027 LQPLPIPTQVWMDISMDFVGGLPKVRGKDTVLVVVDRLTKFAHFYALGHPYSAKDVAVLF 1086
L PLP+P + W D+SMDF+ GLP G T+ VVV+R +K H L ++A VA LF
Sbjct: 426 LCPLPVPQRPWEDLSMDFIVGLPPYHGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASLF 247
Query: 1087 IKELVKLHGFPTTIVSDRDPLFLSNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRCLE 1146
+ ++KLHGFP +IVSDRDPLF+S+FW++LF+++GT L++S++YHPQTDGQTEV NR +E
Sbjct: 246 LNIVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVIE 67
Query: 1147 TYLRCFVGPKPKQWLDWLPWAE 1168
YLR FV +P+ ++PW E
Sbjct: 66 QYLRAFVHGRPRNLGRFIPWVE 1
>BQ299538
Length = 426
Score = 114 bits (286), Expect(2) = 6e-38
Identities = 53/101 (52%), Positives = 70/101 (68%)
Frame = +3
Query: 1116 LFKMAGTQLKLSTSYHPQTDGQTEVTNRCLETYLRCFVGPKPKQWLDWLPWAEFWFNTNY 1175
+ K+ GT LK+STSYHP DGQT N CLET+LRCFV +PK + WL WAE+W+NTN+
Sbjct: 129 IVKLPGTYLKMSTSYHP*IDGQT--VNHCLETFLRCFVADQPKM*VQWLSWAEYWYNTNF 302
Query: 1176 NSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEVNQLFQQRD 1216
++S G +PF+ +YGR PP+L + RVE V + Q RD
Sbjct: 303 HASTGTTPFEVVYGRKPPVLNRFLPGEVRVEAVRRELQDRD 425
Score = 63.2 bits (152), Expect(2) = 6e-38
Identities = 26/44 (59%), Positives = 36/44 (81%)
Frame = +1
Query: 1073 LGHPYSAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWKEL 1116
L HPYSA+ +A +F KE+V LHG P +++SD DP+F+S+FWKEL
Sbjct: 1 LKHPYSARVLAEIFTKEVVHLHGVPASVLSDEDPIFVSSFWKEL 132
>AW830191
Length = 372
Score = 144 bits (362), Expect = 4e-34
Identities = 70/119 (58%), Positives = 84/119 (69%)
Frame = +3
Query: 1149 LRCFVGPKPKQWLDWLPWAEFWFNTNYNSSAGMSPFKALYGRDPPLLLKAGAIPSRVEEV 1208
LRC G KP+QW L WAEFWFNTNYN+S ++PFK LYG DPP LLK I S EEV
Sbjct: 6 LRCLTGTKPQQWPKRLSWAEFWFNTNYNNSLKLTPFKVLYGCDPPHLLKGAIISSTAEEV 185
Query: 1209 NQLFQQRDGVLGELKQNLLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIATR 1267
N + RD +L +LK NL KAQ+QMK AD+ RR + N GDWVYLKLQPY+ +S+A +
Sbjct: 186 NVMTNDRDQMLHDLKGNLAKAQNQMK-YADRSRRSIPLNVGDWVYLKLQPYRQRSLARK 359
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 122 bits (305), Expect = 1e-27
Identities = 75/211 (35%), Positives = 108/211 (50%), Gaps = 19/211 (9%)
Frame = +1
Query: 505 VEKLVAEMLEAGVIRP-SISPFSSPIILVKKKDG------------------GWRLCVDY 545
V K V ++LE G+I P S S + SP+++V KK+G W+LC+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 546 RALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKDADIEKTAFRTHEGH 605
R LN+ T + FP+P ++++L+ L G + LD GY+QI + D EK AF G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 606 YEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVYSPDLLTHTTHLQQVLTL 665
+ + +PFGL NAP+TFQ M + + K VF DD V+ P L + L+ VL
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 666 LADHALKANRKKCSFGQTSLEYLGHIVSGEG 696
+ L N +KC F LGH +S G
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRG 633
>CD487724
Length = 676
Score = 121 bits (304), Expect = 2e-27
Identities = 71/217 (32%), Positives = 110/217 (49%), Gaps = 2/217 (0%)
Frame = +3
Query: 300 IVVLIDCGASHNFISHKLVEQLNLTVAETQTYTVEVGDGYKIRCKGKCSQLGIMIQALEI 359
+V L+D G++HNF+ LV QL L T V VG+G+ ++C C + I IQ +E
Sbjct: 24 LVYLVDGGSTHNFVQQPLVSQLGLPCRSTPPLRVMVGNGHHLKCTTICEAIPISIQNIEF 203
Query: 360 VQNFYLFGLNGMDVVLGIEWLASLGDIKANFGKMELTLKRGKEVYTLKGDPSLTKSELSY 419
+ + Y+ + G ++VLG++WL +LG I ++ + + + LKG+ L++
Sbjct: 204 LVHLYVLPIVGANIVLGVQWLKTLGPILVDYNSLSMQFFYQHRLVQLKGESEAQLGLLNH 383
Query: 420 GALMQILLEEGEGLLVHCD--TGEKVNKKECEIPEIITGVLLQFDTVFQDPQGLPPRRRH 477
L ++ H T +P+ I +L QF +FQ PQGLPP R
Sbjct: 384 HQLRRLHQTHEPVTYFHIAILTENTSPTSSPPLPQPIQHLLDQFSALFQ*PQGLPPARET 563
Query: 478 DHAIHLKEGAQVPNLRPYKCPHYQKNEVEKLVAEMLE 514
DH IHL ++ N+R Y PHY NE+E V ML+
Sbjct: 564 DHHIHLLP*SEPVNMRLY*YPHY*NNEIEHQVNLMLQ 674
>CA820403 weakly similar to GP|13273463|gb| pol protein integrase region
{Ginkgo biloba}, partial (52%)
Length = 421
Score = 94.0 bits (232), Expect(2) = 1e-25
Identities = 47/76 (61%), Positives = 54/76 (70%), Gaps = 1/76 (1%)
Frame = -3
Query: 1086 FIKELVKLHGFPTTIVSDRDPLFL-SNFWKELFKMAGTQLKLSTSYHPQTDGQTEVTNRC 1144
FIKE VKLHG ++IVSD D LFL S FW ELFKM GT+LK S +YHPQ D T+V NRC
Sbjct: 335 FIKEAVKLHGCSSSIVSDWDRLFLIS*FWTELFKMEGTKLKFSLAYHPQPDSHTKVVNRC 156
Query: 1145 LETYLRCFVGPKPKQW 1160
+E L+C K KQW
Sbjct: 155 IEMNLQCLTTSKRKQW 108
Score = 42.7 bits (99), Expect(2) = 1e-25
Identities = 17/34 (50%), Positives = 26/34 (76%)
Frame = -1
Query: 1057 VLVVVDRLTKFAHFYALGHPYSAKDVAVLFIKEL 1090
++V+V R TK+AHF L HPY AK+V+ + +K+L
Sbjct: 421 IMVIVYRFTKYAHFVVLSHPYLAKEVSEVLLKKL 320
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 109 bits (272), Expect = 1e-23
Identities = 72/211 (34%), Positives = 102/211 (48%), Gaps = 19/211 (9%)
Frame = +1
Query: 505 VEKLVAEMLEAGVIRP-SISPFSSPIILVKKKDG------------------GWRLCVDY 545
V K V ++LEAG+I P S S + SP+ +V KK G WR+C+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 546 RALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKDADIEKTAFRTHEGH 605
R LN+ T + +P+P ++++L L + LD SGY+QI + D EKTAF
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 606 YEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVYSPDLLTHTTHLQQVLTL 665
+ + MPFGL NA +TFQ M + + K VF DD + +L++VL
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 666 LADHALKANRKKCSFGQTSLEYLGHIVSGEG 696
L N +KC F LGH +S G
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRG 633
>BI317638 weakly similar to GP|9294238|dbj| contains similarity to reverse
transcriptase~gene_id:K11J14.5 {Arabidopsis thaliana},
partial (5%)
Length = 420
Score = 94.7 bits (234), Expect(2) = 3e-23
Identities = 49/109 (44%), Positives = 66/109 (59%)
Frame = -2
Query: 1009 DCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISMDFVGGLPKVRGKDTVLVVVDRLTKFA 1068
+C CQ +KY T LL PL +P + W D+S+DF+ GL +LVVVD +K
Sbjct: 329 NCLDCQHTKYETKRIVDLLCPLLVPHRPWEDLSLDFITGLLPYHVHTAILVVVDHFSKGI 150
Query: 1069 HFYALGHPYSAKDVAVLFIKELVKLHGFPTTIVSDRDPLFLSNFWKELF 1117
H L ++A VA LFI + KLHG P ++VSD D LF+S+FW+ELF
Sbjct: 149 HLGMLPSSHTAHTVACLFIDSVAKLHGLPRSLVSDCDLLFVSHFWQELF 3
Score = 33.9 bits (76), Expect(2) = 3e-23
Identities = 13/29 (44%), Positives = 20/29 (68%)
Frame = -1
Query: 975 TPTGGHSGHFRTYKRIASFLYWEGMKTDI 1003
T TGGH+G +T ++ +YW GM+TD+
Sbjct: 420 TTTGGHTGIAKTLA*LSKNIYWFGMRTDV 334
>BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 424
Score = 107 bits (267), Expect = 4e-23
Identities = 56/120 (46%), Positives = 78/120 (64%)
Frame = +1
Query: 1226 LLKAQHQMKVQADKHRRLVEFNEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDR 1285
L +AQ M A+ HRR + N GDWVYLK++P++ S+ R KL RFYGPY ++ +
Sbjct: 25 LERAQSLMVKHANNHRRPHDINVGDWVYLKIRPHRQGSMPPRLHPKLTARFYGPYLVMRQ 204
Query: 1286 VGPVAYKLELPAHSKIHPVFHVSLLKQALKPTQQPQPLPPMLNEELELEVVPEDVIDHRE 1345
VG VA++L+LP+ ++IHPVFHVS LK+AL Q + LPP L + EL P ++ RE
Sbjct: 205 VGAVAFQLQLPSEARIHPVFHVSQLKRALGNHQAQEELPPDLEHQAEL-YFPVQILKIRE 381
>TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (5%)
Length = 761
Score = 98.2 bits (243), Expect = 2e-20
Identities = 59/145 (40%), Positives = 84/145 (57%), Gaps = 5/145 (3%)
Frame = +1
Query: 1247 NEGDWVYLKLQPYKLKSIATRPCAKLAPRFYGPYKILDRVGPVAYKLELPAHSKIHPVFH 1306
N GDWV +KL+P++ S + +KL R+YGP+++ +R+G V Y+L+L AHS+IHPVFH
Sbjct: 1 NIGDWVLVKLRPHRQGSASETTYSKLTKRYYGPFEVQERLGKVVYRLKLTAHSRIHPVFH 180
Query: 1307 VSLLKQAL--KPTQQPQPLPPMLNEELELEVVPEDVIDHR---EDAQGNLEVLIKWEQLP 1361
VSLLK + T PLP M E E P VID + D VL++W
Sbjct: 181 VSLLKAFVGDPETTHAGPLPVMRTE--EATNTPLTVIDSKLVPADNGPRRMVLVQWPSAS 354
Query: 1362 TCDNTWELAAVIQDKFPLFPLEDKV 1386
D +WE V++++ + LEDKV
Sbjct: 355 RQDASWEDWQVLRER---YNLEDKV 420
>TC233069
Length = 881
Score = 77.8 bits (190), Expect(2) = 6e-20
Identities = 40/75 (53%), Positives = 50/75 (66%)
Frame = +2
Query: 999 MKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISMDFVGGLPKVRGKDTVL 1058
M D+ + + C CQ++KYST GLLQPLPIP QVW DISMDFV LP GK +
Sbjct: 224 MARDVCDHICACTNCQQNKYSTQK*FGLLQPLPIP*QVWKDISMDFVTHLPPS*GKKMIW 403
Query: 1059 VVVDRLTKFAHFYAL 1073
V+VD TK++HF +L
Sbjct: 404 VIVDCWTKYSHFLSL 448
Score = 39.7 bits (91), Expect(2) = 6e-20
Identities = 23/70 (32%), Positives = 35/70 (49%)
Frame = +1
Query: 928 KVIQDLLISPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISECHDTPTGGHSGHFRTY 987
K++Q L + T+ P + GLL Y L + S +++ ECHD+ GGHSG T
Sbjct: 16 KLLQQKLDNATT-PDCTCQDGLLLYCTHLFIPLESELRAILLGECHDSLLGGHSGIKGTL 192
Query: 988 KRIASFLYWE 997
R+ W+
Sbjct: 193 NRLFVSFAWQ 222
>CF922488
Length = 741
Score = 95.5 bits (236), Expect = 1e-19
Identities = 60/165 (36%), Positives = 87/165 (52%)
Frame = +3
Query: 532 VKKKDGGWRLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKD 591
V K+DG +CVDYR LN + +KFP+P I L+D FS +D SGY+QI++
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 592 ADIEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVYSPD 651
D+EKT F T G + + M FGL N +T+Q M + + K V+ DD++V S
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 652 LLTHTTHLQQVLTLLADHALKANRKKCSFGQTSLEYLGHIVSGEG 696
H +L+++ L + L+ N KC F S + L I S G
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RG 497
>BM084967
Length = 426
Score = 95.1 bits (235), Expect = 2e-19
Identities = 48/137 (35%), Positives = 75/137 (54%)
Frame = -2
Query: 927 VKVIQDLLISPTSHPGFEFKKGLLYYKNRLVLSKASNRIPMIISECHDTPTGGHSGHFRT 986
V++ Q +L PT++P + L+ + L + IP ++ E H +PT H G +T
Sbjct: 416 VQLQQAILAEPTTYPDHSVIQDLILKNGCIWLPSGFSFIPTLLLEYHSSPTDAHIGVTKT 237
Query: 987 YKRIASFLYWEGMKTDIKNFVEDCEICQRSKYSTLAPGGLLQPLPIPTQVWMDISMDFVG 1046
R++ W G++ D++ FV C CQ +KY GLL PLP+P + W D+S +F+
Sbjct: 236 MARLSENFTWIGIRKDVEQFVAACLDCQYTKYEAQKMAGLLCPLPVPCRPWEDLSFNFII 57
Query: 1047 GLPKVRGKDTVLVVVDR 1063
GL + RG +LVVV R
Sbjct: 56 GLSEFRGYTAILVVVGR 6
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 92.0 bits (227), Expect = 2e-18
Identities = 50/142 (35%), Positives = 79/142 (55%)
Frame = +3
Query: 540 RLCVDYRALNKVTIPNKFPIPIIEELLDELGGAKVFSKLDLKSGYHQIRMKDADIEKTAF 599
R+CVDYR LN+ + + FP+P I+ L+ + +FS +D SGY+QI+M D+EKT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 600 RTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLKPYLRKFALVFFDDILVYSPDLLTHTTHL 659
T G + + VM FGL N +T+ M + + + K + D+++ S H +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 660 QQVLTLLADHALKANRKKCSFG 681
Q + L + L+ N +KC FG
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFG 428
>TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 664
Score = 90.5 bits (223), Expect = 5e-18
Identities = 55/137 (40%), Positives = 82/137 (59%), Gaps = 4/137 (2%)
Frame = +2
Query: 1254 LKLQPYKLKSIATRP---CAKLAPRFYGPYKILDRVGPVAYKLELPAHSKIHPVFHVSLL 1310
LKL+P++ +S A P KL+ RF+GP+++++RVG VAY+L+LP +K+HPVFH SLL
Sbjct: 17 LKLRPHR-QSSAKGPEPITGKLSKRFFGPFQVVERVGKVAYRLQLPVDAKLHPVFHCSLL 193
Query: 1311 KQAL-KPTQQPQPLPPMLNEELELEVVPEDVIDHREDAQGNLEVLIKWEQLPTCDNTWEL 1369
K P PLPP L + + + P ++ R ++EVL++W+ L D TWE
Sbjct: 194 KPFQGNPPDTAAPLPPTLFDHQSV-IAPLVILATRTVNDDDIEVLVQWQGLSPDDATWEK 370
Query: 1370 AAVIQDKFPLFPLEDKV 1386
+ + F LEDKV
Sbjct: 371 WTELCKE---FHLEDKV 412
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 66,517,172
Number of Sequences: 63676
Number of extensions: 995265
Number of successful extensions: 4498
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 4419
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4481
length of query: 1430
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1321
effective length of database: 5,698,948
effective search space: 7528310308
effective search space used: 7528310308
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0281a.3


