

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0281a.2
(153 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
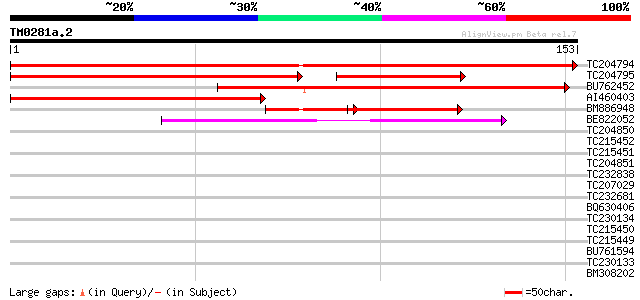
Score E
Sequences producing significant alignments: (bits) Value
TC204794 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete 289 4e-79
TC204795 similar to UP|Q9LWY5 (Q9LWY5) ESTs AU032852(S15362), pa... 159 3e-53
BU762452 155 5e-39
AI460403 134 1e-32
BM886948 53 5e-08
BE822052 similar to GP|4557063|gb|A expressed protein {Arabidops... 41 2e-04
TC204850 similar to GB|AAM16178.1|20334844|AY094022 AT3g12390/T2... 39 0.001
TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%) 39 0.001
TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembl... 39 0.001
TC204851 similar to GB|AAM16178.1|20334844|AY094022 AT3g12390/T2... 38 0.002
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 38 0.002
TC207029 weakly similar to UP|Q7R8E7 (Q7R8E7) Ran-binding protei... 37 0.004
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 37 0.004
BQ630406 37 0.004
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 37 0.004
TC215450 similar to UP|P93488 (P93488) NAP1Ps, partial (30%) 37 0.005
TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%) 37 0.005
BU761594 similar to GP|22654971|gb At1g80930/F23A5_23 {Arabidops... 36 0.006
TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 36 0.006
BM308202 36 0.006
>TC204794 similar to UP|Q9LK52 (Q9LK52) Dbj|BAA90629.1, complete
Length = 1037
Score = 289 bits (739), Expect = 4e-79
Identities = 142/153 (92%), Positives = 150/153 (97%)
Frame = +1
Query: 1 MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLR 60
MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPR++GQDTQDELKRRNLR
Sbjct: 115 MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRKEGQDTQDELKRRNLR 294
Query: 61 DELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSD 120
DELE+RERRHFSSKNKSY DDRDHGK SHLFLEG+KRD+ED IVARSVDADDSDVEVKSD
Sbjct: 295 DELEDRERRHFSSKNKSY-DDRDHGKGSHLFLEGSKRDIEDRIVARSVDADDSDVEVKSD 471
Query: 121 EESDDDDDDEDDTEALLAELEQIKKERAEEKLR 153
++SDDD+DDEDDTEALLAELEQIKKERAEEKLR
Sbjct: 472 DDSDDDEDDEDDTEALLAELEQIKKERAEEKLR 570
>TC204795 similar to UP|Q9LWY5 (Q9LWY5) ESTs AU032852(S15362), partial (49%)
Length = 421
Score = 159 bits (403), Expect(2) = 3e-53
Identities = 75/79 (94%), Positives = 79/79 (99%)
Frame = +1
Query: 1 MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLR 60
MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPR++GQDTQDELKRRNLR
Sbjct: 79 MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRKEGQDTQDELKRRNLR 258
Query: 61 DELEERERRHFSSKNKSYS 79
DELE+RERRHFSSKNKSY+
Sbjct: 259 DELEDRERRHFSSKNKSYA 315
Score = 65.1 bits (157), Expect(2) = 3e-53
Identities = 30/35 (85%), Positives = 34/35 (96%)
Frame = +3
Query: 89 HLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEES 123
HLFLEG+KRD+ED IVARSVDADDSDVEVKSD++S
Sbjct: 315 HLFLEGSKRDIEDRIVARSVDADDSDVEVKSDDDS 419
>BU762452
Length = 422
Score = 155 bits (393), Expect = 5e-39
Identities = 85/140 (60%), Positives = 93/140 (65%), Gaps = 45/140 (32%)
Frame = +2
Query: 57 RNLRDELEERERRHFSSKNKSY-------------------------------------- 78
RNLRDELE+RERRHFSSKNKSY
Sbjct: 2 RNLRDELEDRERRHFSSKNKSYGKVISV*RFSIFLFVAFELHHSCQMDIFSLFLIVICNC 181
Query: 79 -------SDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDED 131
+DDRDHGK SHLFLEG+KR++EDHIV RSVDADDSDVEVKSD++SDDD+DDED
Sbjct: 182 SILFYFLADDRDHGKGSHLFLEGSKREMEDHIVPRSVDADDSDVEVKSDDDSDDDEDDED 361
Query: 132 DTEALLAELEQIKKERAEEK 151
DTEALLAELEQIKKERAEEK
Sbjct: 362 DTEALLAELEQIKKERAEEK 421
>AI460403
Length = 289
Score = 134 bits (338), Expect = 1e-32
Identities = 63/69 (91%), Positives = 67/69 (96%)
Frame = +2
Query: 1 MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLR 60
MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPR++GQDTQDELKRRNLR
Sbjct: 83 MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRKEGQDTQDELKRRNLR 262
Query: 61 DELEERERR 69
D+ + RERR
Sbjct: 263 DDWKNRERR 289
>BM886948
Length = 420
Score = 53.1 bits (126), Expect = 5e-08
Identities = 24/31 (77%), Positives = 29/31 (93%)
Frame = +2
Query: 92 LEGTKRDVEDHIVARSVDADDSDVEVKSDEE 122
L G+KR++EDHIV RSVDADDSDVEVKSD++
Sbjct: 200 LTGSKREMEDHIVPRSVDADDSDVEVKSDDD 292
Score = 49.3 bits (116), Expect = 7e-07
Identities = 23/25 (92%), Positives = 23/25 (92%)
Frame = +3
Query: 70 HFSSKNKSYSDDRDHGKSSHLFLEG 94
HFSSKNKSY DDRDHGK SHLFLEG
Sbjct: 3 HFSSKNKSY-DDRDHGKGSHLFLEG 74
>BE822052 similar to GP|4557063|gb|A expressed protein {Arabidopsis
thaliana}, partial (28%)
Length = 668
Score = 41.2 bits (95), Expect = 2e-04
Identities = 26/93 (27%), Positives = 39/93 (40%)
Frame = -1
Query: 42 KPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVED 101
K D + +DE + D+ +E FS + + +D D D E
Sbjct: 533 KDASDSDEDEDEEDDHDTNDQNDEEGDEDFSGEGEEEADPED--------------DPEA 396
Query: 102 HIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
+ S D DDSD + D + DDD +DED+ E
Sbjct: 395 NGAGGSNDDDDSDEDDDDDNDGDDDGEDEDEKE 297
Score = 26.9 bits (58), Expect = 3.8
Identities = 17/56 (30%), Positives = 25/56 (44%)
Frame = -1
Query: 80 DDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEA 135
DD GK ++ + ED + DD D ++DEE D+D E + EA
Sbjct: 572 DDLCKGKHTYRLNKDASDSDEDED-----EEDDHDTNDQNDEEGDEDFSGEGEEEA 420
>TC204850 similar to GB|AAM16178.1|20334844|AY094022 AT3g12390/T2E22_130
{Arabidopsis thaliana;} , partial (85%)
Length = 878
Score = 38.9 bits (89), Expect = 0.001
Identities = 23/55 (41%), Positives = 32/55 (57%), Gaps = 3/55 (5%)
Frame = +1
Query: 102 HIVARSVDADDSDVEVKSDEESDDDDDDEDD--TEALLAELE-QIKKERAEEKLR 153
H+ + + D+ VE DEE DDDDDDEDD E L + + K+ R+E+K R
Sbjct: 88 HLEQQKIHDDEPVVEDDDDEEVDDDDDDEDDDHDEGLEGDASGRSKQTRSEKKSR 252
>TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%)
Length = 609
Score = 38.5 bits (88), Expect = 0.001
Identities = 14/26 (53%), Positives = 19/26 (72%)
Frame = +3
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTE 134
D DD D+E DE+ +DDDDD+DD +
Sbjct: 144 DEDDEDIEEDDDEDEEDDDDDDDDDD 221
Score = 34.3 bits (77), Expect = 0.024
Identities = 13/44 (29%), Positives = 28/44 (63%)
Frame = +3
Query: 108 VDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
++ D+ D +++ D++ D++DDD+DD + E + KK+ + K
Sbjct: 135 LEDDEDDEDIEEDDDEDEEDDDDDDDDDDDEEESKTKKKSSAPK 266
Score = 34.3 bits (77), Expect = 0.024
Identities = 17/52 (32%), Positives = 29/52 (55%)
Frame = +3
Query: 98 DVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAE 149
D+ED ++ DD + E + D++ DDDDDDE++++ K RA+
Sbjct: 132 DLEDDEDDEDIEEDDDEDE-EDDDDDDDDDDDEEESKTKKKSSAPKKSGRAQ 284
>TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembly protein,
partial (20%)
Length = 777
Score = 38.5 bits (88), Expect = 0.001
Identities = 14/26 (53%), Positives = 19/26 (72%)
Frame = +2
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTE 134
D DD D+E DE+ +DDDDD+DD +
Sbjct: 95 DEDDEDIEEDDDEDEEDDDDDDDDDD 172
Score = 33.5 bits (75), Expect = 0.041
Identities = 13/34 (38%), Positives = 21/34 (61%)
Frame = +2
Query: 98 DVEDHIVARSVDADDSDVEVKSDEESDDDDDDED 131
D+ED ++ DD + E D++ DDDDD+E+
Sbjct: 83 DLEDDEDDEDIEEDDDEDEEDDDDDDDDDDDEEE 184
Score = 33.1 bits (74), Expect = 0.053
Identities = 12/39 (30%), Positives = 26/39 (65%)
Frame = +2
Query: 108 VDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKE 146
++ D+ D +++ D++ D++DDD+DD + E + KK+
Sbjct: 86 LEDDEDDEDIEEDDDEDEEDDDDDDDDDDDEEESKTKKK 202
>TC204851 similar to GB|AAM16178.1|20334844|AY094022 AT3g12390/T2E22_130
{Arabidopsis thaliana;} , partial (86%)
Length = 868
Score = 38.1 bits (87), Expect = 0.002
Identities = 25/76 (32%), Positives = 39/76 (50%), Gaps = 8/76 (10%)
Frame = +2
Query: 86 KSSHLFLEGTKRDVE---DHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELE- 141
KS LF T+ E H+ + + D+ E +DEE DDDDD++D+ + LE
Sbjct: 53 KSKSLFTMTTQTQEELLAQHLEQQKIHDDEPVAEDDNDEEEDDDDDEDDEDDDHADGLEG 232
Query: 142 ----QIKKERAEEKLR 153
+ K+ R+E+K R
Sbjct: 233 DASGRSKQTRSEKKSR 280
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 37.7 bits (86), Expect = 0.002
Identities = 30/113 (26%), Positives = 46/113 (40%), Gaps = 10/113 (8%)
Frame = +2
Query: 46 DGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVA 105
D +D +DE EE + N DD G + + D E+ V
Sbjct: 326 DAEDDEDE----------EEDDDDDEGDDNDDEEDDAPDGGDD----DDDEDDEEEGDVQ 463
Query: 106 RSVDADDSDVEVKSDEESDDDDDDEDD----------TEALLAELEQIKKERA 148
R + DD D + D + D+D+D+ED+ TE L+ LE ++E A
Sbjct: 464 RGGEPDDDDNDSDDDSDDDEDEDEEDEEEQGEEEDLGTEYLIRPLETAEEEEA 622
Score = 35.0 bits (79), Expect = 0.014
Identities = 26/94 (27%), Positives = 40/94 (41%), Gaps = 5/94 (5%)
Frame = +2
Query: 46 DGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDV-----E 100
+G+D + R+E +R + +N + D + L + + +V E
Sbjct: 107 NGRDQRCRATGGKRREEAAQRISKSRGERNVLWFLDSKMKATKELSEQVEENEVKVLVEE 286
Query: 101 DHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
D SVD D E DEE DDDDD+ DD +
Sbjct: 287 DDEEYESVDEVVDDAEDDEDEEEDDDDDEGDDND 388
Score = 35.0 bits (79), Expect = 0.014
Identities = 25/83 (30%), Positives = 39/83 (46%), Gaps = 5/83 (6%)
Frame = +2
Query: 74 KNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSD-----EESDDDDD 128
+++ DD D G + D ED D DD D E + D E DDD+D
Sbjct: 341 EDEEEDDDDDEGDDND--------DEEDDAPDGGDDDDDEDDEEEGDVQRGGEPDDDDND 496
Query: 129 DEDDTEALLAELEQIKKERAEEK 151
+DD++ E E+ ++E+ EE+
Sbjct: 497 SDDDSDDDEDEDEEDEEEQGEEE 565
Score = 32.0 bits (71), Expect = 0.12
Identities = 24/92 (26%), Positives = 41/92 (44%), Gaps = 3/92 (3%)
Frame = +2
Query: 44 RRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHI 103
R G ++E +R + E SK K+ + + + + + + + D E
Sbjct: 128 RATGGKRREEAAQRISKSRGERNVLWFLDSKMKATKELSEQVEENEVKVLVEEDDEEYES 307
Query: 104 VARSVDA---DDSDVEVKSDEESDDDDDDEDD 132
V VD D+ + E D+E DD+DD+EDD
Sbjct: 308 VDEVVDDAEDDEDEEEDDDDDEGDDNDDEEDD 403
Score = 27.3 bits (59), Expect = 2.9
Identities = 27/104 (25%), Positives = 42/104 (39%), Gaps = 13/104 (12%)
Frame = +2
Query: 42 KPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYS----DDRDHGKSSHLFLEGTKR 97
+P D D+ D+ DE +E E+ Y + + ++S F
Sbjct: 473 EPDDDDNDSDDDSDDDEDEDEEDEEEQGEEEDLGTEYLIRPLETAEEEEASSDF------ 634
Query: 98 DVEDHIVARSVDADDSDVEV--------KSD-EESDDDDDDEDD 132
+ E++ D DD D E +SD ++SDDDD EDD
Sbjct: 635 EPEENGEEEEEDVDDEDCEKAEAPPKRKRSDKDDSDDDDGGEDD 766
>TC207029 weakly similar to UP|Q7R8E7 (Q7R8E7) Ran-binding protein, partial
(19%)
Length = 712
Score = 37.0 bits (84), Expect = 0.004
Identities = 27/125 (21%), Positives = 57/125 (45%), Gaps = 14/125 (11%)
Frame = +1
Query: 33 RDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFL 92
+D + T + ++ + DE ++N + + +E+E+ H K + D ++ G+ +
Sbjct: 1 KDEETDTLKEKGKNDEGEDDEENKKNKKKDKKEKEKDH---KKEKKEDGKEEGEKEKSKV 171
Query: 93 EGTKRDVEDHIVARSVDADDSD----VEVKSDEESDDDD----------DDEDDTEALLA 138
E + RD++ + + +D + EVK ++ +D D D D AL
Sbjct: 172 EVSVRDIDIEETTKEGEKEDKEKDDGKEVKEKKKKEDKDKKEKKKVTGKDKTKDLSALKQ 351
Query: 139 ELEQI 143
+LE+I
Sbjct: 352 KLEKI 366
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 37.0 bits (84), Expect = 0.004
Identities = 31/99 (31%), Positives = 45/99 (45%), Gaps = 3/99 (3%)
Frame = +1
Query: 55 KRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSD 114
+R +R R R + S+ DD D+ E + D ED D DD D
Sbjct: 139 RRIRVRG*SSRRRRGR*RRRRLSFDDDDDN--------EDEEDDDEDDAPDGGDDDDDED 294
Query: 115 VEVKSDEE---SDDDDDDEDDTEALLAELEQIKKERAEE 150
E +SD + DDDD++DD E E E+ ++E+ EE
Sbjct: 295 EEEESDVQRGGEPDDDDNDDDDE---DEDEEDEEEQGEE 402
Score = 35.0 bits (79), Expect = 0.014
Identities = 18/54 (33%), Positives = 28/54 (51%)
Frame = +2
Query: 99 VEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKL 152
VE++ V VD ++ + DE DD +DDEDD + LL +K + K+
Sbjct: 95 VEENEVTVLVDEEEDEEYESEDEVVDDGEDDEDDDDYLLTMTTTMKTRKTTMKM 256
Score = 26.2 bits (56), Expect = 6.5
Identities = 16/35 (45%), Positives = 19/35 (53%), Gaps = 9/35 (25%)
Frame = +1
Query: 109 DADDSDVEV--------KSD-EESDDDDDDEDDTE 134
D DD D E +SD ++SDDDD EDD E
Sbjct: 508 DVDDEDGEKSEAPPKRKRSDKDDSDDDDGGEDDDE 612
>BQ630406
Length = 328
Score = 37.0 bits (84), Expect = 0.004
Identities = 28/83 (33%), Positives = 41/83 (48%), Gaps = 5/83 (6%)
Frame = +1
Query: 7 PTWAPAKGGNEQGGTRIFGP--SQKYSSRDIASHTTLKPR-RDGQ--DTQDELKRRNLRD 61
P W P + G G TR+ G +Q++S R+ + +PR R+ + D E+ R RD
Sbjct: 76 PNWRPRRLGGGLGTTRVGGEEVNQRHSGREQQQSRSEEPRVREDRHADRDREISRERGRD 255
Query: 62 ELEERERRHFSSKNKSYSDDRDH 84
+ ERER S + DRDH
Sbjct: 256 KDRERERSREHSHER--VRDRDH 318
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 37.0 bits (84), Expect = 0.004
Identities = 25/99 (25%), Positives = 44/99 (44%), Gaps = 4/99 (4%)
Frame = +2
Query: 40 TLKPRRDGQDTQDELKRRNLRD----ELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGT 95
T + +D DT+D+ ++ D + ++ + FS + D D ++ E
Sbjct: 248 TSEENKDASDTEDDDDDDDVNDGEDGDDDDEDDEDFSGDDGGEEADSDDDPEANGGGESD 427
Query: 96 KRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
D +D D DD D + +E DDD++DED+ E
Sbjct: 428 DDDEDD-------DDDDDDNDEDDGDEDDDDEEDEDEDE 523
Score = 33.9 bits (76), Expect = 0.031
Identities = 14/37 (37%), Positives = 22/37 (58%)
Frame = +2
Query: 96 KRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDD 132
K E++ A + DD D +V E+ DDDD+D++D
Sbjct: 242 KHTSEENKDASDTEDDDDDDDVNDGEDGDDDDEDDED 352
>TC215450 similar to UP|P93488 (P93488) NAP1Ps, partial (30%)
Length = 672
Score = 36.6 bits (83), Expect = 0.005
Identities = 17/29 (58%), Positives = 19/29 (64%), Gaps = 3/29 (10%)
Frame = +3
Query: 109 DADDSDVEVKSDEESD---DDDDDEDDTE 134
D DD D+E DEE + DDDDDEDD E
Sbjct: 216 DEDDEDIEEDDDEEEEEDEDDDDDEDDEE 302
Score = 34.3 bits (77), Expect = 0.024
Identities = 15/37 (40%), Positives = 25/37 (67%)
Frame = +3
Query: 98 DVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
D+ED ++ DD D E + DE+ DDD+DDE++++
Sbjct: 204 DLEDDEDDEDIEEDD-DEEEEEDEDDDDDEDDEEESK 311
>TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%)
Length = 1495
Score = 36.6 bits (83), Expect = 0.005
Identities = 13/26 (50%), Positives = 19/26 (73%)
Frame = +2
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTE 134
D DD D+E DEE ++D+DD+DD +
Sbjct: 1049 DEDDEDIEEDDDEEEEEDEDDDDDED 1126
Score = 35.0 bits (79), Expect = 0.014
Identities = 18/52 (34%), Positives = 29/52 (55%)
Frame = +2
Query: 98 DVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAE 149
D+ED ++ DD D E + DE+ DDD+DDE++++ K RA+
Sbjct: 1037 DLEDDEDDEDIEEDD-DEEEEEDEDDDDDEDDEEESKTKKKSSAPKKSGRAQ 1189
>BU761594 similar to GP|22654971|gb At1g80930/F23A5_23 {Arabidopsis
thaliana}, partial (13%)
Length = 397
Score = 36.2 bits (82), Expect = 0.006
Identities = 24/57 (42%), Positives = 34/57 (59%), Gaps = 1/57 (1%)
Frame = +1
Query: 91 FLEGTKRDVEDHIVARSVDADDS-DVEVKSDEESDDDDDDEDDTEALLAELEQIKKE 146
FLE KR E + +S+ ++S D E D ESDDDDD++D+++ E QIK E
Sbjct: 136 FLENEKRYEE---LKKSMLGEESEDDEEGLDSESDDDDDEDDESDEEEEEQMQIKDE 297
Score = 28.5 bits (62), Expect = 1.3
Identities = 24/107 (22%), Positives = 49/107 (45%), Gaps = 2/107 (1%)
Frame = +1
Query: 38 HTTLKPRRDGQDTQDELKRR-NLRDELE-ERERRHFSSKNKSYSDDRDHGKSSHLFLEGT 95
+ ++P D + +D++ +L +E++ E F +++ + + L
Sbjct: 13 YPAVRPELDLVEQEDQITHEVSLDEEIDPEISLDIFKPDPNFLENEKRYEELKKSMLGEE 192
Query: 96 KRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQ 142
D E+ + + S D DD D +SDEE ++ +D+TE L L +
Sbjct: 193 SEDDEEGLDSESDDDDDED--DESDEEEEEQMQIKDETETNLVNLRR 327
>TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 850
Score = 36.2 bits (82), Expect = 0.006
Identities = 25/99 (25%), Positives = 44/99 (44%), Gaps = 3/99 (3%)
Frame = +3
Query: 40 TLKPRRDGQDTQDELKRRNLRD---ELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTK 96
T + +D DT+D+ N+ D + ++ E FS + D D ++
Sbjct: 252 TSEENKDASDTEDDDDDDNVNDGEDDNDDEEDEDFSGDDGGEEADSDDDPEANGGGGSDD 431
Query: 97 RDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEA 135
D +D D DD + E DE+ +D+ DD++D E+
Sbjct: 432 DDGDD-------DEDDDNDEDDGDEDDEDEGDDDEDEES 527
Score = 30.0 bits (66), Expect = 0.45
Identities = 13/36 (36%), Positives = 20/36 (55%)
Frame = +3
Query: 96 KRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDED 131
K E++ A + DD D V E+ +DD++DED
Sbjct: 246 KHTSEENKDASDTEDDDDDDNVNDGEDDNDDEEDED 353
>BM308202
Length = 433
Score = 36.2 bits (82), Expect = 0.006
Identities = 23/59 (38%), Positives = 32/59 (53%), Gaps = 1/59 (1%)
Frame = +2
Query: 80 DDRDHGKSSHLFLE-GTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALL 137
DD+D +S E G +R +D D+D D E S+EE +DDDDD+ D + LL
Sbjct: 176 DDQDEMYASARIYEIGRRRPTDD-------DSDPDDAE--SEEEDEDDDDDDPDVDPLL 325
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.306 0.127 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,612,146
Number of Sequences: 63676
Number of extensions: 51594
Number of successful extensions: 2023
Number of sequences better than 10.0: 268
Number of HSP's better than 10.0 without gapping: 796
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1301
length of query: 153
length of database: 12,639,632
effective HSP length: 89
effective length of query: 64
effective length of database: 6,972,468
effective search space: 446237952
effective search space used: 446237952
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0281a.2


