

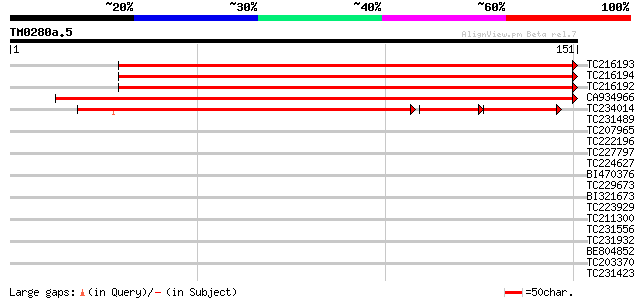
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0280a.5
(151 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216193 wound-induced protein 191 1e-49
TC216194 similar to UP|Q6KF83 (Q6KF83) Pseudo-hevein (Fragment),... 188 1e-48
TC216192 similar to UP|Q6KF83 (Q6KF83) Pseudo-hevein (Fragment),... 187 2e-48
CA934966 similar to GP|3511147|gb| PR-4 type protein {Vitis vini... 186 3e-48
TC234014 similar to UP|PR4A_TOBAC (P29062) Pathogenesis-related ... 144 1e-46
TC231489 similar to GB|AAN73294.1|25141199|BT002297 At5g12010/F1... 30 0.34
TC207965 homologue to UP|DAPA_SOYBN (Q42800) Dihydrodipicolinate... 28 2.2
TC222196 weakly similar to UP|Q77MR7 (Q77MR7) UL45 (Envelope tra... 27 2.8
TC227797 similar to UP|Q8H1N8 (Q8H1N8) At1g20880/F9H16_14, parti... 27 3.7
TC224627 similar to UP|PSBY_ARATH (O49347) Photosystem II core c... 27 3.7
BI470376 weakly similar to GP|14090338|dbj P0638D12.5 {Oryza sat... 27 4.8
TC229673 similar to GB|AAM26654.1|20856224|AY101533 At1g14810/F1... 27 4.8
BI321673 27 4.8
TC223929 27 4.8
TC211300 UP|Q8CH69 (Q8CH69) Survival of motor neuron protein (Fr... 26 6.3
TC231556 similar to UP|Q8GUH2 (Q8GUH2) Expressed protein (At1g01... 26 6.3
TC231932 similar to GB|AAS75312.1|45680886|AY568529 multidomain ... 26 6.3
BE804852 26 8.3
TC203370 weakly similar to UP|Q9LX02 (Q9LX02) ESTs AU082316(E336... 26 8.3
TC231423 similar to UP|Q9LID4 (Q9LID4) Similarity to disease res... 26 8.3
>TC216193 wound-induced protein
Length = 741
Score = 191 bits (484), Expect = 1e-49
Identities = 82/122 (67%), Positives = 98/122 (80%)
Frame = +1
Query: 30 GQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLAWRKKYGWTAFCGPAGAHGRD 89
G+SA+NV ATY+ Y P+Q GW+L AYC+TW+A+KP +WR KYGWTAFCGP G GRD
Sbjct: 226 GESASNVRATYHYYEPEQHGWDLNAVSAYCSTWDASKPYSWRSKYGWTAFCGPVGPRGRD 405
Query: 90 SCGKCLRVRNTATGDEATVRIVDQCANGGLDLDVNVFRRLDSNGQGNHQGHLTVNYRFVN 149
SCGKCLRV NT TG VRIVDQC+NGGLDLDV VF R+D++G+G QGHL VNY+FV+
Sbjct: 406 SCGKCLRVTNTGTGANTIVRIVDQCSNGGLDLDVGVFNRIDTDGRGYQQGHLIVNYQFVD 585
Query: 150 CG 151
CG
Sbjct: 586 CG 591
>TC216194 similar to UP|Q6KF83 (Q6KF83) Pseudo-hevein (Fragment), partial
(91%)
Length = 694
Score = 188 bits (477), Expect = 1e-48
Identities = 82/122 (67%), Positives = 97/122 (79%)
Frame = +2
Query: 30 GQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLAWRKKYGWTAFCGPAGAHGRD 89
G+SA+NV ATY+ Y P+Q GW+L AYC+TW+A KP +WR KYGWTAFCGP G GRD
Sbjct: 146 GESASNVRATYHNYLPEQHGWDLNAVSAYCSTWDAAKPYSWRSKYGWTAFCGPVGPRGRD 325
Query: 90 SCGKCLRVRNTATGDEATVRIVDQCANGGLDLDVNVFRRLDSNGQGNHQGHLTVNYRFVN 149
+CGKCLRV NT TG VRIVDQC+NGGLDLDV VF R+D++G+G QGHL VNY+FVN
Sbjct: 326 ACGKCLRVTNTGTGANIIVRIVDQCSNGGLDLDVGVFNRIDTDGRGYQQGHLIVNYQFVN 505
Query: 150 CG 151
CG
Sbjct: 506 CG 511
>TC216192 similar to UP|Q6KF83 (Q6KF83) Pseudo-hevein (Fragment), partial
(85%)
Length = 654
Score = 187 bits (474), Expect = 2e-48
Identities = 81/122 (66%), Positives = 97/122 (79%)
Frame = +1
Query: 30 GQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLAWRKKYGWTAFCGPAGAHGRD 89
G+SA+NV ATY+ Y P+Q GW+L AYC+TW+A+KP +WR KYGWTAFCGP G GRD
Sbjct: 118 GESASNVRATYHYYEPEQHGWDLNAVSAYCSTWDASKPYSWRSKYGWTAFCGPVGPRGRD 297
Query: 90 SCGKCLRVRNTATGDEATVRIVDQCANGGLDLDVNVFRRLDSNGQGNHQGHLTVNYRFVN 149
SCGKCLRV NT TG VRIVDQC+NGGLDLDV VF +D++G+G QGHL VNY+FV+
Sbjct: 298 SCGKCLRVINTGTGANTIVRIVDQCSNGGLDLDVGVFNWIDTDGRGYQQGHLIVNYQFVD 477
Query: 150 CG 151
CG
Sbjct: 478 CG 483
>CA934966 similar to GP|3511147|gb| PR-4 type protein {Vitis vinifera},
partial (88%)
Length = 451
Score = 186 bits (473), Expect = 3e-48
Identities = 83/139 (59%), Positives = 104/139 (74%)
Frame = +1
Query: 13 KLSIVAVWWLIAATFANGQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLAWRK 72
K+S+ V + A+ QSA NV ATY+LY P+Q W+LL AYC+TW+A+K LAWR
Sbjct: 4 KVSLFVVCVVCILALASAQSATNVRATYHLYQPEQHNWDLLAVSAYCSTWDADKSLAWRS 183
Query: 73 KYGWTAFCGPAGAHGRDSCGKCLRVRNTATGDEATVRIVDQCANGGLDLDVNVFRRLDSN 132
KYGWTAFCGP+G G+ SCG+CLRV NT T + TVRIVDQC+NGGLDLD+ F++LD++
Sbjct: 184 KYGWTAFCGPSGPQGQQSCGRCLRVTNTRTNAQETVRIVDQCSNGGLDLDIGPFQKLDTD 363
Query: 133 GQGNHQGHLTVNYRFVNCG 151
G G QGHL VNY FV+CG
Sbjct: 364 GNGIAQGHLIVNYDFVDCG 420
>TC234014 similar to UP|PR4A_TOBAC (P29062) Pathogenesis-related protein
PR-4A precursor, partial (78%)
Length = 559
Score = 144 bits (362), Expect(3) = 1e-46
Identities = 65/96 (67%), Positives = 73/96 (75%), Gaps = 6/96 (6%)
Frame = +2
Query: 19 VWWLIAAT------FANGQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNANKPLAWRK 72
VW ++ T +G+SA NV ATY+LYNP+QIGWNL+TA AYCATW+ANKPLAWRK
Sbjct: 50 VWCVMMVTRRRTRVLVSGESAKNVRATYHLYNPEQIGWNLVTASAYCATWDANKPLAWRK 229
Query: 73 KYGWTAFCGPAGAHGRDSCGKCLRVRNTATGDEATV 108
KY WTAFCGP G G SCGKCLRV NT TG EATV
Sbjct: 230 KYAWTAFCGPVGPRGXKSCGKCLRVTNTGTGTEATV 337
Score = 40.0 bits (92), Expect(3) = 1e-46
Identities = 16/21 (76%), Positives = 19/21 (90%)
Frame = +1
Query: 127 RRLDSNGQGNHQGHLTVNYRF 147
R+LD+NG GN QGHLT+NYRF
Sbjct: 394 RQLDTNGVGNQQGHLTLNYRF 456
Score = 39.3 bits (90), Expect(3) = 1e-46
Identities = 17/17 (100%), Positives = 17/17 (100%)
Frame = +3
Query: 110 IVDQCANGGLDLDVNVF 126
IVDQCANGGLDLDVNVF
Sbjct: 342 IVDQCANGGLDLDVNVF 392
>TC231489 similar to GB|AAN73294.1|25141199|BT002297 At5g12010/F14F18_180
{Arabidopsis thaliana;} , partial (22%)
Length = 634
Score = 30.4 bits (67), Expect = 0.34
Identities = 20/56 (35%), Positives = 25/56 (43%), Gaps = 1/56 (1%)
Frame = -2
Query: 49 GWNLLTAGAYCATWNANKPLAWRKKYGWTAFCGP-AGAHGRDSCGKCLRVRNTATG 103
GW L GA+C++W P A R W CG AG R S G C + +G
Sbjct: 486 GWLL---GAWCSSW*RRSPGAGRSCRRWPCSCGSGAGIPLRGSPGCCTAATSPWSG 328
>TC207965 homologue to UP|DAPA_SOYBN (Q42800) Dihydrodipicolinate synthase,
chloroplast precursor (DHDPS) , partial (98%)
Length = 1291
Score = 27.7 bits (60), Expect = 2.2
Identities = 19/55 (34%), Positives = 24/55 (43%), Gaps = 8/55 (14%)
Frame = +3
Query: 55 AGAYCATWNA----NKPLAWRKKYGWTAF----CGPAGAHGRDSCGKCLRVRNTA 101
AG C WNA +KPL W+ GW A C G H C R R+++
Sbjct: 516 AGFCC--WNACCPSHKPLLWQNLLGWYACSLSKCAFHGTHNNLQCACTDRSRHSS 674
>TC222196 weakly similar to UP|Q77MR7 (Q77MR7) UL45 (Envelope transmembrane
protein), partial (11%)
Length = 880
Score = 27.3 bits (59), Expect = 2.8
Identities = 17/62 (27%), Positives = 25/62 (39%), Gaps = 5/62 (8%)
Frame = +2
Query: 43 YNPQQIGWNLLTAGAYCAT-----WNANKPLAWRKKYGWTAFCGPAGAHGRDSCGKCLRV 97
++P+ G NL + A +T W + L Y C A HG SC +R
Sbjct: 275 FHPRATGHNLGPSNAAHSTVVGLGWQRTRNLMSPSLYEGPCLCSRARIHGGSSCSSIMRR 454
Query: 98 RN 99
R+
Sbjct: 455 RS 460
>TC227797 similar to UP|Q8H1N8 (Q8H1N8) At1g20880/F9H16_14, partial (59%)
Length = 1737
Score = 26.9 bits (58), Expect = 3.7
Identities = 12/26 (46%), Positives = 15/26 (57%)
Frame = -2
Query: 89 DSCGKCLRVRNTATGDEATVRIVDQC 114
DS G+C +VR T T E +VD C
Sbjct: 785 DSVGRCTQVRMTYTHKEGKPILVDSC 708
>TC224627 similar to UP|PSBY_ARATH (O49347) Photosystem II core complex
proteins psbY, chloroplast precursor (L-arginine
metabolising enzyme) (L-AME) [Contains: Photosystem II
protein psbY-1 (psbY-A1); Photosystem II protein psbY-2
(psbY-A2)], partial (49%)
Length = 1049
Score = 26.9 bits (58), Expect = 3.7
Identities = 14/34 (41%), Positives = 16/34 (46%)
Frame = +2
Query: 69 AWRKKYGWTAFCGPAGAHGRDSCGKCLRVRNTAT 102
AWR GW C G SCG C R + TA+
Sbjct: 494 AWR--LGWVGGCAACVGGGF*SCGCCERQQGTAS 589
>BI470376 weakly similar to GP|14090338|dbj P0638D12.5 {Oryza sativa
(japonica cultivar-group)}, partial (5%)
Length = 429
Score = 26.6 bits (57), Expect = 4.8
Identities = 17/68 (25%), Positives = 28/68 (41%), Gaps = 2/68 (2%)
Frame = -3
Query: 9 ITQRKLSIVAVWWLIAATFANGQSANNVMATYNLYNPQQIGWNLLTAGAYCATWNAN--K 66
+ ++ V W I F A N ++ ++ G N T+ +CATW + K
Sbjct: 376 VPDNEIGAVESEWSIG*CFY*KVEARNAYISFEVW-----GGNYYTSRCFCATWYSCGWK 212
Query: 67 PLAWRKKY 74
W+ KY
Sbjct: 211 TFNWKYKY 188
>TC229673 similar to GB|AAM26654.1|20856224|AY101533 At1g14810/F10B6_6
{Arabidopsis thaliana;} , partial (51%)
Length = 1442
Score = 26.6 bits (57), Expect = 4.8
Identities = 12/30 (40%), Positives = 17/30 (56%)
Frame = -1
Query: 69 AWRKKYGWTAFCGPAGAHGRDSCGKCLRVR 98
+WR++ W A+ GP G R CG RV+
Sbjct: 875 SWRRRRRW-AWGGPGGGPRRGGCGAKQRVK 789
>BI321673
Length = 318
Score = 26.6 bits (57), Expect = 4.8
Identities = 13/38 (34%), Positives = 16/38 (41%)
Frame = +1
Query: 59 CATWNANKPLAWRKKYGWTAFCGPAGAHGRDSCGKCLR 96
C W++ K WR+ GWTA P C K R
Sbjct: 52 CRKWSSKK--TWRRCRGWTAARCPRATTTPFRCSKAAR 159
>TC223929
Length = 567
Score = 26.6 bits (57), Expect = 4.8
Identities = 14/31 (45%), Positives = 16/31 (51%), Gaps = 2/31 (6%)
Frame = +2
Query: 53 LTAG--AYCATWNANKPLAWRKKYGWTAFCG 81
L AG A+ ATW +KP G AFCG
Sbjct: 185 LAAGTAAWAATWKLSKPFRINLSAGAGAFCG 277
>TC211300 UP|Q8CH69 (Q8CH69) Survival of motor neuron protein (Fragment),
partial (5%)
Length = 1151
Score = 26.2 bits (56), Expect = 6.3
Identities = 17/65 (26%), Positives = 27/65 (41%), Gaps = 7/65 (10%)
Frame = +1
Query: 40 YNLYNPQQIG--WNLL-----TAGAYCATWNANKPLAWRKKYGWTAFCGPAGAHGRDSCG 92
Y P IG WN + T G+Y +TW W+ + CG + R C
Sbjct: 142 YTKQIPFPIGSTWNFVILTQDTLGSY*STWIVRMRSLWKHIDPCSMNCGNFRSKKRCLCA 321
Query: 93 KCLRV 97
+C+++
Sbjct: 322 RCMKL 336
>TC231556 similar to UP|Q8GUH2 (Q8GUH2) Expressed protein (At1g01560),
partial (16%)
Length = 456
Score = 26.2 bits (56), Expect = 6.3
Identities = 9/30 (30%), Positives = 15/30 (50%)
Frame = +1
Query: 47 QIGWNLLTAGAYCATWNANKPLAWRKKYGW 76
Q GW + +C +W N+P R+ + W
Sbjct: 136 QRGWTTIMVQCWC*SWCWNRPWDVRRAWDW 225
>TC231932 similar to GB|AAS75312.1|45680886|AY568529 multidomain cyclophilin
type peptidyl-prolyl cis-trans isomerase {Arabidopsis
thaliana;} , partial (13%)
Length = 329
Score = 26.2 bits (56), Expect = 6.3
Identities = 10/28 (35%), Positives = 18/28 (63%)
Frame = +1
Query: 124 NVFRRLDSNGQGNHQGHLTVNYRFVNCG 151
NV ++++ G + +GH TV + +NCG
Sbjct: 226 NVLKKIEEFG--DEEGHPTVTVKIINCG 303
>BE804852
Length = 238
Score = 25.8 bits (55), Expect = 8.3
Identities = 16/47 (34%), Positives = 20/47 (42%)
Frame = +2
Query: 62 WNANKPLAWRKKYGWTAFCGPAGAHGRDSCGKCLRVRNTATGDEATV 108
WNA AW C H D C +CL + +AT +E TV
Sbjct: 44 WNAVSLWAWDIAVDNRTICLN---HIMDLCIECLAYQTSATSEECTV 175
>TC203370 weakly similar to UP|Q9LX02 (Q9LX02) ESTs AU082316(E3368), partial
(27%)
Length = 1507
Score = 25.8 bits (55), Expect = 8.3
Identities = 9/20 (45%), Positives = 13/20 (65%)
Frame = +1
Query: 73 KYGWTAFCGPAGAHGRDSCG 92
K+ W A+CG +GA+ CG
Sbjct: 691 KWEWIAYCGDSGANWS*RCG 750
>TC231423 similar to UP|Q9LID4 (Q9LID4) Similarity to disease resistance
response protein, partial (40%)
Length = 650
Score = 25.8 bits (55), Expect = 8.3
Identities = 14/40 (35%), Positives = 21/40 (52%), Gaps = 3/40 (7%)
Frame = -2
Query: 45 PQQIGWNLLTAGAYC---ATWNANKPLAWRKKYGWTAFCG 81
P+ +G +L AGA A W +W+KK+ W +F G
Sbjct: 295 PKSVG-DLSVAGAAATRTAVWFWPLTTSWKKKWRWLSFSG 179
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.134 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,126,258
Number of Sequences: 63676
Number of extensions: 121009
Number of successful extensions: 547
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 538
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 544
length of query: 151
length of database: 12,639,632
effective HSP length: 89
effective length of query: 62
effective length of database: 6,972,468
effective search space: 432293016
effective search space used: 432293016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0280a.5


