

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0276.6
(483 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
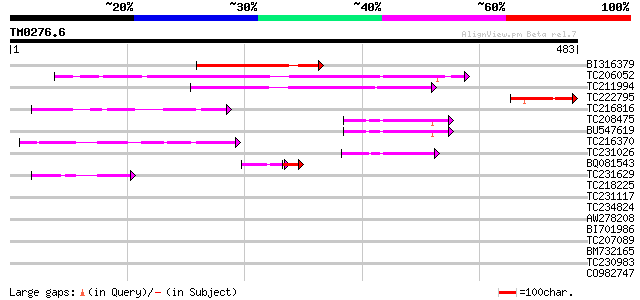
Score E
Sequences producing significant alignments: (bits) Value
BI316379 166 2e-41
TC206052 similar to UP|Q6GUE8 (Q6GUE8) NIM1-like protein 1, part... 156 2e-38
TC211994 weakly similar to UP|Q6GUF2 (Q6GUF2) NIM1-like protein ... 103 2e-22
TC222795 UP|Q76PT8 (Q76PT8) D5L protein, partial (18%) 74 1e-13
TC216816 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like pro... 54 2e-07
TC208475 similar to UP|AKR_ARATH (Q05753) Ankyrin repeat protein... 49 7e-06
BU547619 47 1e-05
TC216370 46 3e-05
TC231026 similar to UP|Q9SQK3 (Q9SQK3) Ankyrin repeat protein EM... 46 4e-05
BQ081543 32 3e-04
TC231629 42 8e-04
TC218225 weakly similar to UP|IPP_HUMAN (Q9Y573) Actin-binding p... 41 0.001
TC231117 similar to UP|Q7QH40 (Q7QH40) AgCP8016 (Fragment), part... 40 0.002
TC234824 similar to UP|Q9I7K2 (Q9I7K2) CG4655-PA, partial (4%) 40 0.002
AW278208 39 0.004
BI701986 39 0.004
TC207089 39 0.004
BM732165 homologue to SP|Q9TV66|HCN4 Potassium/sodium hyperpolar... 39 0.007
TC230983 similar to GB|AAB61117.1|2194142|F20P5 ESTs gb|N38288,g... 38 0.009
CO982747 38 0.012
>BI316379
Length = 305
Score = 166 bits (420), Expect = 2e-41
Identities = 84/108 (77%), Positives = 93/108 (85%)
Frame = +2
Query: 160 SMVEKASIEDVMKVLLASRKQDMHQLWTTCSHLVAKSGLPPEVLAKHLPIDIVAKIEELR 219
SMVEKASI+DVMKVL+ASRKQ+M QLW+TCSHLVAKSGLPPEVLAKHLPID+VAKIEELR
Sbjct: 2 SMVEKASIDDVMKVLIASRKQEMQQLWSTCSHLVAKSGLPPEVLAKHLPIDVVAKIEELR 181
Query: 220 LKSTLARRSLIPHHHHHHHHHGHHDMGAAADLEDQKIRRMRRALDSSD 267
LKS+LARRSL+P HH HHH +EDQKI+RMRRALDSSD
Sbjct: 182 LKSSLARRSLLPGHHQHHH-------DLTPGMEDQKIQRMRRALDSSD 304
>TC206052 similar to UP|Q6GUE8 (Q6GUE8) NIM1-like protein 1, partial (79%)
Length = 1757
Score = 156 bits (395), Expect = 2e-38
Identities = 113/355 (31%), Positives = 176/355 (48%), Gaps = 2/355 (0%)
Frame = +3
Query: 39 HRCILAARSLFFRKFFCGPDPPPPSGNLDSPGGPRVNSPRPGGVIPVNSVGYEVFLLMLQ 98
HRCILA+RS FF + F G+ + G + N ++P VGYE FL+ L
Sbjct: 18 HRCILASRSKFFHELF-----KREKGSSEKEGKLKYNM---NDLLPYGKVGYEAFLIFLG 173
Query: 99 FLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAVDLALDTLAAARYFGVEQLALLTQKQL 158
++Y+G++ P + C + C H C A++ A++ + A+ F + +L L Q++L
Sbjct: 174 YVYTGKLKPSPMEVS---TCVDNVCAHDACRPAINFAVELMYASSIFQIPELVSLFQRRL 344
Query: 159 ASMVEKASIEDVMKVLLASRKQDMHQLWTTCSHLVAKSGLPPEVLAKHLPIDIVAKIEEL 218
+ + KA +EDV+ +L + +QL C VA+S L + + LP ++ K++ L
Sbjct: 345 LNFIGKALVEDVIPILTVAFHCQSNQLVNQCIDRVARSDLDQISIDQELPHELSQKVKLL 524
Query: 219 RLKSTLARRSLIPHHHHHHHHHGHHDMGAAADLEDQKIRRMRRALDSSDVELVKLMVMGE 278
R K +D L ++I R+ +ALDS DVELVKL++
Sbjct: 525 RRKP---------------QQDVENDASVVDALSLKRITRIHKALDSDDVELVKLLLNES 659
Query: 279 GLNLDEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVL 338
+ LDEA AL YA C +VV +L LG A+VN + G T LHIAA P ++ L
Sbjct: 660 DITLDEANALHYAAAYCDPKVVSEVLGLGLANVNLRNS-RGYTVLHIAAMRKEPSIIVSL 836
Query: 339 LDHHADPNVRTVDGVTPLDILRTLT--SDFLFKGAVPGLTHIEPNKLRLCLELVQ 391
L A + T DG + + I R LT D+ K E NK R+C+++++
Sbjct: 837 LTKGACASDLTFDGQSAVSICRRLTRPKDYHAKTE----QGKETNKDRICIDVLE 989
>TC211994 weakly similar to UP|Q6GUF2 (Q6GUF2) NIM1-like protein 1, partial
(33%)
Length = 630
Score = 103 bits (257), Expect = 2e-22
Identities = 71/209 (33%), Positives = 106/209 (49%)
Frame = +2
Query: 155 QKQLASMVEKASIEDVMKVLLASRKQDMHQLWTTCSHLVAKSGLPPEVLAKHLPIDIVAK 214
Q+ L + VEKA +EDV+ +L+A+ + QL + C VA+S L K LP ++V +
Sbjct: 2 QRHLLNFVEKALVEDVIPILMAAFNCQLDQLLSQCIRRVARSDFDNTSLEKELPREVVTE 181
Query: 215 IEELRLKSTLARRSLIPHHHHHHHHHGHHDMGAAADLEDQKIRRMRRALDSSDVELVKLM 274
I+ LRL + L ++ IRR+ +ALDS DVEL+KL+
Sbjct: 182 IKLLRLP---------------FQPESTPNAMEVESLNEKSIRRIHKALDSDDVELLKLL 316
Query: 275 VMGEGLNLDEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDM 334
+ + LD+A AL YA +V++ +L LG AD+ G T LH+AA P +
Sbjct: 317 LNESSVTLDDAHALHYACAYSDSKVIQEVLSLGMADI-LRRNSRGYTVLHVAARRKDPSI 493
Query: 335 VAVLLDHHADPNVRTVDGVTPLDILRTLT 363
+ LL+ A + T DG T L I + LT
Sbjct: 494 LVALLNKGACASDTTPDGQTALAICQRLT 580
>TC222795 UP|Q76PT8 (Q76PT8) D5L protein, partial (18%)
Length = 693
Score = 74.3 bits (181), Expect = 1e-13
Identities = 41/68 (60%), Positives = 48/68 (70%), Gaps = 11/68 (16%)
Frame = +3
Query: 427 NEDHNSSSSNA----------NLNLDSRLVYLNLGA-AQMSGDDDSSHGSHREAMNQAMY 475
+EDHN++SS++ NLNLDSRLVYLNLGA A MS D++ REAMNQ MY
Sbjct: 6 SEDHNNNSSSSSGNNNNNNIGNLNLDSRLVYLNLGATASMSSRLDNAE-DEREAMNQTMY 182
Query: 476 HHHHSHDY 483
HHHHSHDY
Sbjct: 183 HHHHSHDY 206
>TC216816 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like protein X,
partial (5%)
Length = 1610
Score = 53.9 bits (128), Expect = 2e-07
Identities = 50/171 (29%), Positives = 73/171 (42%)
Frame = +1
Query: 19 INGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDSPGGPRVNSPR 78
+N SDVTF VEG+ +AHR L A S FR F GG R R
Sbjct: 703 VNNVTLSDVTFLVEGKRFYAHRICLLASSDAFRAMF--------------DGGYREKEAR 840
Query: 79 PGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHCTSAVDLALDT 138
I + ++ +EVF LM++F+Y+G V I ++D+A D
Sbjct: 841 D---IEIPNIRWEVFELMMRFIYTGSVDI-----------------------SLDIAQDL 942
Query: 139 LAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWTTC 189
L AA + +E L L + +A + S+E+V + S + L TC
Sbjct: 943 LRAADQYLLEGLKRLCEYTIA---QDISLENVSSMYELSEAFNAISLRHTC 1086
>TC208475 similar to UP|AKR_ARATH (Q05753) Ankyrin repeat protein,
chloroplast precursor (AKRP), partial (51%)
Length = 1057
Score = 48.5 bits (114), Expect = 7e-06
Identities = 36/101 (35%), Positives = 50/101 (48%), Gaps = 7/101 (6%)
Frame = +2
Query: 285 ALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHAD 344
A + YAV S + +K LL L D+N G TPLH+A + D+V +LL AD
Sbjct: 500 ATLMHYAVLTASTQTIKILL-LYNVDINLQDN-YGWTPLHLAVQAQRTDLVRLLLIKGAD 673
Query: 345 PNVRTVDGVTPLDI-------LRTLTSDFLFKGAVPGLTHI 378
++ DG+TPLD+ RT LFK L+H+
Sbjct: 674 KTLKNEDGLTPLDLCLYNGQSARTYELIKLFKQPQRRLSHV 796
>BU547619
Length = 406
Score = 47.4 bits (111), Expect = 1e-05
Identities = 37/101 (36%), Positives = 49/101 (47%), Gaps = 7/101 (6%)
Frame = -1
Query: 285 ALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHHAD 344
A + AV S + VK LL L D+N P G TPLH+A + D+V +LL AD
Sbjct: 391 ATLMXXAVLTXSTQTVKILL-LYNVDINLPDN-YGWTPLHLAVQAQRTDLVRLLLIKGAD 218
Query: 345 PNVRTVDGVTPLDI-------LRTLTSDFLFKGAVPGLTHI 378
++ DG+TPLD+ RT LFK L H+
Sbjct: 217 KTLKNEDGLTPLDLCLYNGQCARTYELIKLFKQPQRRLRHV 95
>TC216370
Length = 1349
Score = 46.2 bits (108), Expect = 3e-05
Identities = 50/188 (26%), Positives = 78/188 (40%)
Frame = +3
Query: 9 SLSLDYLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFRKFFCGPDPPPPSGNLDS 68
S+ + +LL +G+ SDV F V + AH+ +LAARS FR GP
Sbjct: 348 SIGQKFGHLLESGKG-SDVNFEVNDDIFAAHKLVLAARSPVFRAQLFGP----------- 491
Query: 69 PGGPRVNSPRPGGVIPVNSVGYEVFLLMLQFLYSGQVSIVPQKHEPRPNCGERACWHTHC 128
+ I V + VF +L F+Y + P+ E ++
Sbjct: 492 ------MKDQNTQCIKVEDMEAPVFKALLHFIY----------WDSLPDMQELTGLNSKW 623
Query: 129 TSAVDLALDTLAAARYFGVEQLALLTQKQLASMVEKASIEDVMKVLLASRKQDMHQLWTT 188
S + +A LAAA +G+E+L L+ + AS+ E +I V L + + QL
Sbjct: 624 ASTL-MAQHLLAAADRYGLERLRLMCE---ASLCEDVAINTVATTLALAEQHHCFQLKAV 791
Query: 189 CSHLVAKS 196
C +A S
Sbjct: 792 CLKFIATS 815
>TC231026 similar to UP|Q9SQK3 (Q9SQK3) Ankyrin repeat protein EMB506,
partial (33%)
Length = 750
Score = 45.8 bits (107), Expect = 4e-05
Identities = 32/84 (38%), Positives = 44/84 (52%)
Frame = +3
Query: 283 DEALALPYAVENCSREVVKALLELGAADVNYPSGPSGKTPLHIAAEMVSPDMVAVLLDHH 342
D A L YAV+ ++ VK L++ ADVN G TPLHIA + + D+ +LL +
Sbjct: 42 DGAAPLHYAVQVGAKMTVKLLIKY-KADVNVEDN-EGWTPLHIAIQSRNRDIAKILLVNG 215
Query: 343 ADPNVRTVDGVTPLDILRTLTSDF 366
AD + DG T LD+ DF
Sbjct: 216 ADKTRKNKDGKTALDLSLCYGKDF 287
>BQ081543
Length = 424
Score = 32.3 bits (72), Expect(2) = 3e-04
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = +2
Query: 233 HHHHHHHHGHHDMGAAAD 250
HHHHHHHH G A++
Sbjct: 359 HHHHHHHHRGRQQGKASE 412
Score = 32.0 bits (71), Expect = 0.63
Identities = 11/21 (52%), Positives = 12/21 (56%)
Frame = +2
Query: 232 HHHHHHHHHGHHDMGAAADLE 252
HHHHHHHH G A+ E
Sbjct: 359 HHHHHHHHRGRQQGKASEGSE 421
Score = 29.6 bits (65), Expect(2) = 3e-04
Identities = 14/41 (34%), Positives = 22/41 (53%)
Frame = +2
Query: 198 LPPEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHHHHH 238
LPPE ++ ++E+ +T + R+ HHHHHHH
Sbjct: 266 LPPETNPLAGAGPLLRMLQEMA--TTKSSRNTCHHHHHHHH 382
>TC231629
Length = 1074
Score = 41.6 bits (96), Expect = 8e-04
Identities = 29/90 (32%), Positives = 44/90 (48%), Gaps = 1/90 (1%)
Frame = +2
Query: 19 INGQAFSDVTFSVEGRLVHAHR-CILAARSLFFRKFFCGPDPPPPSGNLDSPGGPRVNSP 77
+N SDVTF VEGR +AHR C+L+ S FR F G
Sbjct: 200 VNNPKLSDVTFLVEGRSFYAHRDCLLS--SDIFRAMFDGS-----------------YRE 322
Query: 78 RPGGVIPVNSVGYEVFLLMLQFLYSGQVSI 107
R I + ++ ++VF LM++++Y+G V +
Sbjct: 323 REAKSIVIPNIKWDVFELMMRYIYTGTVDV 412
>TC218225 weakly similar to UP|IPP_HUMAN (Q9Y573) Actin-binding protein IPP
(MIPP protein), partial (6%)
Length = 1029
Score = 40.8 bits (94), Expect = 0.001
Identities = 21/44 (47%), Positives = 28/44 (62%), Gaps = 1/44 (2%)
Frame = +3
Query: 14 YLNLLINGQAFSDVTFSVEGRLVHAHRCILAARSLFFR-KFFCG 56
+ +L+ + SDVTFSV G HAH+ +LAARS F +FF G
Sbjct: 657 HFGMLLENEEGSDVTFSVGGERFHAHKLVLAARSTAFETEFFNG 788
>TC231117 similar to UP|Q7QH40 (Q7QH40) AgCP8016 (Fragment), partial (6%)
Length = 805
Score = 40.4 bits (93), Expect = 0.002
Identities = 12/13 (92%), Positives = 12/13 (92%)
Frame = +2
Query: 231 PHHHHHHHHHGHH 243
PHHHHHHHHH HH
Sbjct: 212 PHHHHHHHHHHHH 250
Score = 38.5 bits (88), Expect = 0.007
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 239 HHHHHHHHHSHH 274
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 224 HHHHHHHHHHHH 259
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 221 HHHHHHHHHHHH 256
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 230 HHHHHHHHHHHH 265
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 227 HHHHHHHHHHHH 262
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 218 HHHHHHHHHHHH 253
Score = 36.6 bits (83), Expect = 0.026
Identities = 11/13 (84%), Positives = 11/13 (84%)
Frame = +2
Query: 231 PHHHHHHHHHGHH 243
PH HHHHHHH HH
Sbjct: 206 PHPHHHHHHHHHH 244
Score = 34.3 bits (77), Expect = 0.13
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHH H HH
Sbjct: 245 HHHHHHHSHHHH 280
Score = 34.3 bits (77), Expect = 0.13
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHH HH
Sbjct: 242 HHHHHHHHSHHH 277
Score = 34.3 bits (77), Expect = 0.13
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH H
Sbjct: 236 HHHHHHHHHHSH 271
Score = 32.7 bits (73), Expect = 0.37
Identities = 10/13 (76%), Positives = 10/13 (76%)
Frame = +2
Query: 231 PHHHHHHHHHGHH 243
PH H HHHHH HH
Sbjct: 200 PHPHPHHHHHHHH 238
Score = 32.0 bits (71), Expect = 0.63
Identities = 9/11 (81%), Positives = 9/11 (81%)
Frame = +2
Query: 232 HHHHHHHHHGH 242
HHHH HHHH H
Sbjct: 254 HHHHSHHHHAH 286
Score = 30.4 bits (67), Expect = 1.8
Identities = 9/12 (75%), Positives = 9/12 (75%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHH HHH H
Sbjct: 251 HHHHHSHHHHAH 286
>TC234824 similar to UP|Q9I7K2 (Q9I7K2) CG4655-PA, partial (4%)
Length = 426
Score = 40.0 bits (92), Expect = 0.002
Identities = 13/18 (72%), Positives = 14/18 (77%)
Frame = +3
Query: 226 RRSLIPHHHHHHHHHGHH 243
R S+I HHHHH HHH HH
Sbjct: 336 RHSMIKHHHHHRHHHHHH 389
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 375 HHHHHHHHHHHH 410
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 378 HHHHHHHHHHHH 413
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 381 HHHHHHHHHHHH 416
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = +3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 372 HHHHHHHHHHHH 407
>AW278208
Length = 396
Score = 39.3 bits (90), Expect = 0.004
Identities = 11/15 (73%), Positives = 14/15 (93%)
Frame = +2
Query: 229 LIPHHHHHHHHHGHH 243
++PH+HHHHHHH HH
Sbjct: 5 VVPHNHHHHHHHHHH 49
Score = 34.7 bits (78), Expect = 0.097
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +2
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH H
Sbjct: 20 HHHHHHHHHHRH 55
Score = 33.1 bits (74), Expect = 0.28
Identities = 10/15 (66%), Positives = 12/15 (79%)
Frame = +2
Query: 229 LIPHHHHHHHHHGHH 243
L+ H+HHHHHH HH
Sbjct: 2 LVVPHNHHHHHHHHH 46
Score = 32.0 bits (71), Expect = 0.63
Identities = 10/19 (52%), Positives = 11/19 (57%)
Frame = +2
Query: 232 HHHHHHHHHGHHDMGAAAD 250
HHHHHHH H H +D
Sbjct: 29 HHHHHHHRHRRHRNNNCSD 85
Score = 30.4 bits (67), Expect = 1.8
Identities = 9/20 (45%), Positives = 12/20 (60%)
Frame = +2
Query: 232 HHHHHHHHHGHHDMGAAADL 251
HHHHHH H H + + D+
Sbjct: 32 HHHHHHRHRRHRNNNCSDDV 91
>BI701986
Length = 422
Score = 39.3 bits (90), Expect = 0.004
Identities = 16/29 (55%), Positives = 18/29 (61%)
Frame = +2
Query: 212 VAKIEELRLKSTLARRSLIPHHHHHHHHH 240
+A L L S L R +I HHHHHHHHH
Sbjct: 5 IAGFFSLGLSSLLLPRLMIIHHHHHHHHH 91
Score = 35.8 bits (81), Expect = 0.044
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = +2
Query: 224 LARRSLIPHHHHHHHHH 240
L R +I HHHHHHHHH
Sbjct: 44 LPRLMIIHHHHHHHHHH 94
Score = 30.0 bits (66), Expect = 2.4
Identities = 15/27 (55%), Positives = 17/27 (62%), Gaps = 2/27 (7%)
Frame = +1
Query: 57 PDPPPPSGN--LDSPGGPRVNSPRPGG 81
P PPPP N LDSP P + P+PGG
Sbjct: 79 PPPPPPKRNPVLDSPSTP--SPPKPGG 153
>TC207089
Length = 522
Score = 39.3 bits (90), Expect = 0.004
Identities = 11/14 (78%), Positives = 13/14 (92%)
Frame = +1
Query: 230 IPHHHHHHHHHGHH 243
+P+HHHHHHHH HH
Sbjct: 277 VPYHHHHHHHHHHH 318
Score = 38.9 bits (89), Expect = 0.005
Identities = 12/15 (80%), Positives = 12/15 (80%)
Frame = +1
Query: 232 HHHHHHHHHGHHDMG 246
HHHHHHHHH HH G
Sbjct: 286 HHHHHHHHHHHHRAG 330
Score = 35.4 bits (80), Expect = 0.057
Identities = 12/20 (60%), Positives = 12/20 (60%)
Frame = +1
Query: 232 HHHHHHHHHGHHDMGAAADL 251
HHHHHHHHH H G L
Sbjct: 289 HHHHHHHHHHHRAGGLICHL 348
Score = 31.2 bits (69), Expect = 1.1
Identities = 15/39 (38%), Positives = 19/39 (48%), Gaps = 3/39 (7%)
Frame = +1
Query: 216 EELRLKSTLARRSLIPHH---HHHHHHHGHHDMGAAADL 251
EE + +A +L H +HHHHHH HH A L
Sbjct: 220 EEEKPSMCIAMENLTLHQNVPYHHHHHHHHHHHHRAGGL 336
Score = 28.1 bits (61), Expect = 9.1
Identities = 11/27 (40%), Positives = 13/27 (47%)
Frame = +1
Query: 215 IEELRLKSTLARRSLIPHHHHHHHHHG 241
+E L L + HHHHHHH G
Sbjct: 250 MENLTLHQNVPYHHHHHHHHHHHHRAG 330
>BM732165 homologue to SP|Q9TV66|HCN4 Potassium/sodium
hyperpolarization-activated cyclic nucleotide-gated
channel 4, partial (2%)
Length = 423
Score = 38.5 bits (88), Expect = 0.007
Identities = 14/21 (66%), Positives = 15/21 (70%)
Frame = +1
Query: 222 STLARRSLIPHHHHHHHHHGH 242
S L R +I HHHHHHHHH H
Sbjct: 1 SLLLPRLMIIHHHHHHHHHHH 63
Score = 34.7 bits (78), Expect = 0.097
Identities = 10/11 (90%), Positives = 10/11 (90%)
Frame = +1
Query: 233 HHHHHHHHGHH 243
HHHHHHHH HH
Sbjct: 31 HHHHHHHHHHH 63
Score = 30.0 bits (66), Expect = 2.4
Identities = 15/27 (55%), Positives = 17/27 (62%), Gaps = 2/27 (7%)
Frame = +3
Query: 57 PDPPPPSGN--LDSPGGPRVNSPRPGG 81
P PPPP N LDSP P + P+PGG
Sbjct: 48 PPPPPPKRNPVLDSPSTP--SPPKPGG 122
>TC230983 similar to GB|AAB61117.1|2194142|F20P5 ESTs
gb|N38288,gb|T43486,gb|AA395242 come from this gene.
{Arabidopsis thaliana;} , partial (34%)
Length = 938
Score = 38.1 bits (87), Expect = 0.009
Identities = 16/54 (29%), Positives = 26/54 (47%)
Frame = -1
Query: 191 HLVAKSGLPPEVLAKHLPIDIVAKIEELRLKSTLARRSLIPHHHHHHHHHGHHD 244
HL K ++ I ++ + L + L ++ +HHHHHHHH HH+
Sbjct: 632 HLNKKKQSTERIMKNPNSISLLPNLYCTLLANNLRTTNVPKNHHHHHHHHHHHN 471
>CO982747
Length = 196
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = -3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 194 HHHHHHHHHHHH 159
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = -3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 188 HHHHHHHHHHHH 153
Score = 37.7 bits (86), Expect = 0.012
Identities = 11/12 (91%), Positives = 11/12 (91%)
Frame = -3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHHH HH
Sbjct: 191 HHHHHHHHHHHH 156
Score = 30.0 bits (66), Expect = 2.4
Identities = 9/12 (75%), Positives = 9/12 (75%)
Frame = -3
Query: 232 HHHHHHHHHGHH 243
HHHHHHHH H
Sbjct: 176 HHHHHHHHVSCH 141
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,547,183
Number of Sequences: 63676
Number of extensions: 452200
Number of successful extensions: 14672
Number of sequences better than 10.0: 407
Number of HSP's better than 10.0 without gapping: 6039
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10613
length of query: 483
length of database: 12,639,632
effective HSP length: 101
effective length of query: 382
effective length of database: 6,208,356
effective search space: 2371591992
effective search space used: 2371591992
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0276.6


