

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0269b.9
(1085 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
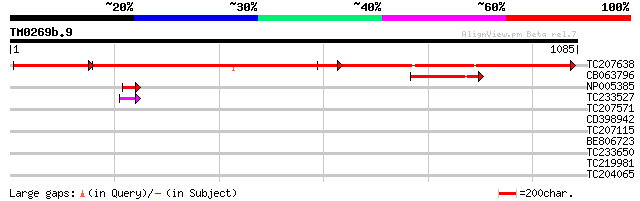
TC207638 UP|RPC2_SOYBN (Q8HVY3) DNA-directed RNA polymerase beta... 721 0.0
CB063796 GP|15778189|g rpoC2 {Glycine max}, partial (6%) 176 4e-44
NP005385 DNA-directed RNA polymerase 46 8e-05
TC233527 weakly similar to UP|Q9LVH0 (Q9LVH0) DNA-directed RNA p... 43 7e-04
TC207571 weakly similar to UP|O65470 (O65470) Serine/threonine k... 39 0.012
CD398942 weakly similar to GP|21592666|gb| receptor-like protein... 33 0.89
TC207115 31 3.4
BE806723 30 4.4
TC233650 similar to UP|Q84JC9 (Q84JC9) BZIP transcription factor... 30 5.8
TC219981 similar to UP|Q8LBQ5 (Q8LBQ5) Receptor-like protein kin... 30 5.8
TC204065 homologue to UP|Q39835 (Q39835) Extensin, partial (29%) 30 7.5
>TC207638 UP|RPC2_SOYBN (Q8HVY3) DNA-directed RNA polymerase beta'' chain
(PEP) (Plastid-encoded RNA polymerase beta'' subunit)
(RNA polymerase beta'' subunit) , complete
Length = 4156
Score = 721 bits (1862), Expect(2) = 0.0
Identities = 364/507 (71%), Positives = 414/507 (80%), Gaps = 28/507 (5%)
Frame = +1
Query: 159 IYIGSRCIVVRNQDIGIGLINRFINFQTQPIFIRTPFTCRNTSWICRLCYGRSPIHGDLV 218
IY GSRCI VRNQDIGIGLINRF QTQPI IRTPFTCRNTSWICRLCYG+SP HG LV
Sbjct: 769 IYRGSRCIAVRNQDIGIGLINRFKTLQTQPISIRTPFTCRNTSWICRLCYGQSPTHGHLV 948
Query: 219 ELGEAVGIIAGQSIGEPGTQLTLITFHTGGVFTGGTAEYVRSPSNGKIKFNENSAYPTRT 278
ELGEAVGIIAGQSIGEPGTQLTL TFHTGGVFTGGTAE VR+P NGKI+FNE+ +PTRT
Sbjct: 949 ELGEAVGIIAGQSIGEPGTQLTLRTFHTGGVFTGGTAEQVRAPYNGKIQFNEDLVHPTRT 1128
Query: 279 RHGHPAFLCYIDLYVTIESNDIMHNVIIPPKSFLLVQNDQYVKSEQIIAEIRAGTYTLNL 338
RHGHPAFLCYIDLYVTIES DI+HNV IPPKSFLLVQN+QYVKSEQ+IAEIRAGTYT NL
Sbjct: 1129 RHGHPAFLCYIDLYVTIESGDIIHNVTIPPKSFLLVQNNQYVKSEQVIAEIRAGTYTFNL 1308
Query: 339 KEKIRKHIFSDSEGEMHWSTNIYHVSEFAYSNVHILPKTSHLWILSGNSHKSDTVSLSLL 398
KEK+RKH++SD EGEMHWST++YH SEF YSNVHILPKTSHLWILSG S++S +VS S+
Sbjct: 1309 KEKVRKHVYSDLEGEMHWSTDVYHASEFRYSNVHILPKTSHLWILSGKSYRSGSVSFSIR 1488
Query: 399 KDQDQMSTHSLPTAKRNTSNFLVSNNQVR-------------------------LCPDHC 433
KDQDQM+ H L T +R+ N L S +VR + DHC
Sbjct: 1489 KDQDQMNIHYLSTGERDFCNLLASKKKVRHKLFRFNPSDKKERRISDYSIFNQIISIDHC 1668
Query: 434 HFMHPTISPDTSNLLAKKRRNRFIIPFLFRSIRERNNELM--PDISVEIPIDGIIHKNSI 491
HF HPTI DT++LLAK+RRNR IIPF F+SI+ER+ ELM IS+EIPI+GI +NSI
Sbjct: 1669 HFTHPTIFHDTTDLLAKRRRNRLIIPFQFQSIQERDKELMLASSISIEIPINGIFRRNSI 1848
Query: 492 LAYFDDPQYRTQSSGIAKYKTIGIHSIFQKED-LIEYRGIREFKPKYQIKVDRFFFIPQE 550
LAYFDDPQYRTQSSGI KY+TIGI+SIF+KED LIEY+G++EFK KYQIKVD+FFFIP+E
Sbjct: 1849 LAYFDDPQYRTQSSGITKYRTIGINSIFKKEDFLIEYQGVKEFKTKYQIKVDQFFFIPEE 2028
Query: 551 VHILSESSSIMVRNNSIIGVNTPITLNKKSRVGGLVRVEKNKKKIELKIFSGDIHFPGEI 610
VHIL ESSSIMVRNNSI+ V+T ITLN +SRV GLVR+EK KKKIELKIFSGDI+FPGE+
Sbjct: 2029 VHILPESSSIMVRNNSIVEVDTLITLNIRSRVRGLVRLEKKKKKIELKIFSGDIYFPGEM 2208
Query: 611 DKISQHSAILIPPEMVKKKILRNQKKK 637
DKIS+H + + KKK R + +K
Sbjct: 2209 DKISRHXCRVDTTKNGKKKFKRKKNEK 2289
Score = 679 bits (1752), Expect = 0.0
Identities = 350/497 (70%), Positives = 399/497 (79%), Gaps = 3/497 (0%)
Frame = +2
Query: 590 KNKKKIELKIFSGDIHFPGEIDKISQHSAILIPPEMVKKKILRNQKKKTNWRYIQWITTT 649
+ KK++ K F G FP + + SA+LIPP VKK +KK NW Y+QWIT T
Sbjct: 2147 RKKKRLN*KYFPGISIFPEKWIRYPDTSAVLIPPRTVKKN--SKEKKMKNWIYVQWITIT 2320
Query: 650 KKKYFVLVRPVILYDIADSINLVKLFPQDLFKEWDNLELKVLNFILYGNGKSIRGILDTS 709
KKKYFVLVRPVILY+IAD INLVKLFPQD+F+E DNLELKV+N+IL GNGKSIRGI DTS
Sbjct: 2321 KKKYFVLVRPVILYEIADRINLVKLFPQDMFQERDNLELKVINYILSGNGKSIRGISDTS 2500
Query: 710 IQLVRTCLVLNWNEDEKSSSIEEALASFVEVSTNGLIRYFLRIDLVKSHISYIRKRNDPS 769
IQLVRTCLVLNW++D+K SSIE A ASFVE+S GL+RYFLRIDLVKSHISYI KRNDPS
Sbjct: 2501 IQLVRTCLVLNWDQDKKFSSIENAHASFVEISIKGLVRYFLRIDLVKSHISYISKRNDPS 2680
Query: 770 SSGLISYNESDRININPFFSIY---KENIQQSLSQKHGTIRMLLNRNKENRSFIILSSSN 826
S SD N+NPF+SIY K +Q+S +Q GT+R LL NKE + F+ILSSSN
Sbjct: 2681 SE---KKEGSDHTNMNPFYSIYIYPKTKLQKSFNQNQGTVRTLLGINKECQFFLILSSSN 2851
Query: 827 CFQMGPFNNVKYHNGIKEEINQFKRNHKIPIKISLGPLGVAPQIANFFSFYHLITHNKIS 886
CFQ+ FN+ KY+NGIKE INQ +R+ IPIK SLGPLG+APQ+A+F FY LITHN+IS
Sbjct: 2852 CFQIRSFNHGKYYNGIKEGINQIQRDAMIPIKNSLGPLGIAPQVAHF-DFYRLITHNQIS 3028
Query: 887 SIKKNLQLNKFKETFQVIKYYLMDENERIYKPDLYNNIILNPFHLNWDFIHPNYCEKTFP 946
IK N QL+K KETFQV +YY +DENERIYKPDL +NIILNPF+LNW F+H NYCEKTF
Sbjct: 3029 IIK-NGQLDKLKETFQVFQYYFLDENERIYKPDLCSNIILNPFYLNWHFLHHNYCEKTFT 3205
Query: 947 IISLGQFICENVCIVQTKNGPNLKSGQVITVQMDFVGIRLANPYLATPGATIHGHYGEML 1006
I+SLGQFICENVCIVQTKN P+LKSGQ++TVQMD VGIR ANPYLATPGAT+HGHYGE+L
Sbjct: 3206 IMSLGQFICENVCIVQTKNAPHLKSGQILTVQMDSVGIRSANPYLATPGATVHGHYGEIL 3385
Query: 1007 YEGDILVTFIYEKSRSGDITQGLPKVEQVLEVRSIDSISMNLEKRIDAWNERITGILGIP 1066
EGDILVTFIYEKSRSGDITQGLPKVEQVLEVRSIDSISMNLEKR+D WN RIT ILGIP
Sbjct: 3386 SEGDILVTFIYEKSRSGDITQGLPKVEQVLEVRSIDSISMNLEKRVDTWNGRITKILGIP 3565
Query: 1067 WSRKISMSRKLLQDNIS 1083
W I + Q IS
Sbjct: 3566 WGFLIGAELTIAQSRIS 3616
Score = 231 bits (589), Expect(2) = 0.0
Identities = 115/152 (75%), Positives = 123/152 (80%)
Frame = +3
Query: 8 DMVCYK*IFATRNESQF*YD*PL*SSPYNVFFGS*GKCISSAPISRYERINVGSPGPND* 67
DMVCYK*IF+TRNES F* D PL*SSPYNV FGS* KC+SS PIS YERINVGS ND*
Sbjct: 318 DMVCYK*IFSTRNES*F*DDRPL*SSPYNVLFGS*RKCVSSTPISGYERINVGSTRTND* 497
Query: 68 FTHSKQFT*RTFFNRIYYFLLWSPQGGCGYCCTNIRCWISYTETC*SGSTHCCTPNRLRY 127
FT+SKQF R FFNRIY FLL S GC YCCTNIRCWISYT+TC*SGSTHCCT N L Y
Sbjct: 498 FTYSKQFARRAFFNRIYNFLLRSS*RGCRYCCTNIRCWISYTQTC*SGSTHCCTSNGLWY 677
Query: 128 RSRDFCKYSE*NDVRKNFDTNINWSCISRRYI 159
S++F +Y + ND KN DTNINWSCISRRYI
Sbjct: 678 HSKNFSEYPKWNDAGKNMDTNINWSCISRRYI 773
>CB063796 GP|15778189|g rpoC2 {Glycine max}, partial (6%)
Length = 422
Score = 176 bits (447), Expect = 4e-44
Identities = 96/142 (67%), Positives = 112/142 (78%), Gaps = 1/142 (0%)
Frame = +3
Query: 767 DPSSSGLISYNESDRININPFFSI-YKENIQQSLSQKHGTIRMLLNRNKENRSFIILSSS 825
+P S I NESDR INPFFSI +E IQQSLS+ HGTIRMLLNRN++ RS IILSSS
Sbjct: 3 NPPGSRFILDNESDRTTINPFFSIDLREKIQQSLSKNHGTIRMLLNRNEKCRSLIILSSS 182
Query: 826 NCFQMGPFNNVKYHNGIKEEINQFKRNHKIPIKISLGPLGVAPQIANFFSFYHLITHNKI 885
NCFQ+ FN+ KY+NGIKE INQ +R+ IPIK SLGPLG+APQ+A+ F FY LITHN+I
Sbjct: 183 NCFQIRSFNHGKYYNGIKEGINQIQRDAMIPIKNSLGPLGIAPQVAH-FDFYRLITHNQI 359
Query: 886 SSIKKNLQLNKFKETFQVIKYY 907
S I KN QL+K KETFQV +YY
Sbjct: 360 SII-KNGQLDKLKETFQVFQYY 422
>NP005385 DNA-directed RNA polymerase
Length = 2934
Score = 46.2 bits (108), Expect = 8e-05
Identities = 21/33 (63%), Positives = 25/33 (75%)
Frame = +1
Query: 217 LVELGEAVGIIAGQSIGEPGTQLTLITFHTGGV 249
LV GE +G++A QSIGEP TQ+TL TFH GV
Sbjct: 802 LVVPGEMIGVVAAQSIGEPATQMTLNTFHFAGV 900
>TC233527 weakly similar to UP|Q9LVH0 (Q9LVH0) DNA-directed RNA polymerase II
largest chain, partial (10%)
Length = 609
Score = 43.1 bits (100), Expect = 7e-04
Identities = 21/39 (53%), Positives = 23/39 (58%)
Frame = +1
Query: 211 SPIHGDLVELGEAVGIIAGQSIGEPGTQLTLITFHTGGV 249
S H +E G VG SIGEPGTQ+TL TFH GV
Sbjct: 172 SRYHSKKMEAGAPVGATGAHSIGEPGTQMTLKTFHFAGV 288
>TC207571 weakly similar to UP|O65470 (O65470) Serine/threonine kinase-like
protein , partial (23%)
Length = 1117
Score = 38.9 bits (89), Expect = 0.012
Identities = 18/22 (81%), Positives = 19/22 (85%)
Frame = +1
Query: 231 SIGEPGTQLTLITFHTGGVFTG 252
SIGEPGTQLTL TF+ G VFTG
Sbjct: 1051 SIGEPGTQLTLRTFYIGKVFTG 1116
>CD398942 weakly similar to GP|21592666|gb| receptor-like protein kinase-like
protein {Arabidopsis thaliana}, partial (7%)
Length = 652
Score = 32.7 bits (73), Expect = 0.89
Identities = 27/84 (32%), Positives = 39/84 (46%), Gaps = 4/84 (4%)
Frame = +1
Query: 660 VILYDIADSINL--VKLFPQDLFKEWDNLELKVLNFILYGNGKSIRGILDTSIQLVRTCL 717
++L I+ S+NL + P+D+F W L+ F+ YGN R L RTC
Sbjct: 250 ILLSPISCSLNLRCILKVPKDIFNGWSAP*LRRYTFVCYGNHN**R--------LFRTCS 405
Query: 718 VLNW--NEDEKSSSIEEALASFVE 739
+ NW N +IE L + VE
Sbjct: 406 LQNWVHNLFPTILTIERCLQNGVE 477
>TC207115
Length = 1182
Score = 30.8 bits (68), Expect = 3.4
Identities = 21/65 (32%), Positives = 29/65 (44%), Gaps = 9/65 (13%)
Frame = +2
Query: 81 NRIYYFLLWSPQGGC--GYCCTNIRCWISYTETC*SGSTHCCTPNRLRYR-------SRD 131
+RI F +W+ + GYC + CW YT C S S+ C R RY +
Sbjct: 257 SRILIFNIWAARSCILFGYCRVDSCCWHIYTLWCISLSSQC----RCRYMCCHGWLGAEP 424
Query: 132 FCKYS 136
+C YS
Sbjct: 425 YCTYS 439
>BE806723
Length = 389
Score = 30.4 bits (67), Expect = 4.4
Identities = 11/32 (34%), Positives = 18/32 (55%)
Frame = -3
Query: 536 KYQIKVDRFFFIPQEVHILSESSSIMVRNNSI 567
++++ + FFF QE H L S +RN +I
Sbjct: 96 RFEVSISHFFFFQQE*HTLCTQGSYFIRNQTI 1
>TC233650 similar to UP|Q84JC9 (Q84JC9) BZIP transcription factor, partial
(17%)
Length = 792
Score = 30.0 bits (66), Expect = 5.8
Identities = 24/107 (22%), Positives = 49/107 (45%), Gaps = 2/107 (1%)
Frame = -1
Query: 623 PEMVKKKILRNQKKKTNWRYIQWITTTKKKYFVLVRPVILYDIA--DSINLVKLFPQDLF 680
P ++K +++R ++ W + + T + + ++ + L LF + +F
Sbjct: 570 PFLLKMEMMRFYTERPFWEWEGYQRRTSSTATLSIVFLVWNSLPLNGQFQLPSLFYKPVF 391
Query: 681 KEWDNLELKVLNFILYGNGKSIRGILDTSIQLVRTCLVLNWNEDEKS 727
W E ++L+F+LY +G + G S +L R C W + E+S
Sbjct: 390 *LWT--EPRILDFLLYAHGVDLAGWFS*SRRLRRKC----WRQKEES 268
>TC219981 similar to UP|Q8LBQ5 (Q8LBQ5) Receptor-like protein kinase-like
protein, partial (19%)
Length = 772
Score = 30.0 bits (66), Expect = 5.8
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 2/64 (3%)
Frame = -2
Query: 660 VILYDIADSINL--VKLFPQDLFKEWDNLELKVLNFILYGNGKSIRGILDTSIQLVRTCL 717
++L I+ S+NL + P+D+F W L+ F+ YGN R L R C
Sbjct: 579 ILLSPISCSLNLRCILKVPKDIFNGWSAP*LRRYTFVCYGNHN**R--------LFRACS 424
Query: 718 VLNW 721
+ NW
Sbjct: 423 LHNW 412
>TC204065 homologue to UP|Q39835 (Q39835) Extensin, partial (29%)
Length = 603
Score = 29.6 bits (65), Expect = 7.5
Identities = 28/98 (28%), Positives = 48/98 (48%), Gaps = 6/98 (6%)
Frame = +3
Query: 357 STNIYHVSEFAYSNVHILPKTSHLWILSGNSH---KSDTVSLSLLKDQDQMSTHSLPTAK 413
+T++ H +F +N+ +LP ++HL +L S +S SLL Q ++ SL +
Sbjct: 30 NTSLPH-PQFTSTNLPLLPISTHLLLLPLTSTLHLHHQYISTSLLLHHPQSTSTSLHHPQ 206
Query: 414 RNTSNFLVSNNQVRLCPDH---CHFMHPTISPDTSNLL 448
++N L+ + L H H HP + +TS LL
Sbjct: 207 FTSTNLLLHPISIHLLHHHHTSTHHHHPQFT-NTSLLL 317
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.142 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,374,993
Number of Sequences: 63676
Number of extensions: 858893
Number of successful extensions: 5785
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 5605
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5770
length of query: 1085
length of database: 12,639,632
effective HSP length: 107
effective length of query: 978
effective length of database: 5,826,300
effective search space: 5698121400
effective search space used: 5698121400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0269b.9


