

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0269b.10
(382 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
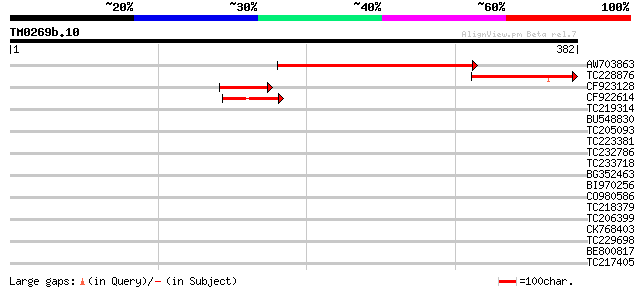
AW703863 256 1e-68
TC228876 similar to UP|YC2A_ATRBE (Q8S8V2) Protein ycf2, partial... 113 1e-25
CF923128 63 3e-10
CF922614 42 5e-04
TC219314 weakly similar to UP|Q7XUE7 (Q7XUE7) OJ991113_30.24 pro... 33 0.002
BU548830 32 0.004
TC205093 31 1.1
TC223381 31 1.1
TC232786 similar to UP|Q947G7 (Q947G7) Thermal hysteresis protei... 30 2.4
TC233718 similar to UP|Q9LTK1 (Q9LTK1) Arabidopsis thaliana geno... 30 2.4
BG352463 similar to GP|11245494|gb| SPATULA {Arabidopsis thalian... 29 4.0
BI970256 29 4.0
CO980586 29 4.0
TC218379 similar to UP|Q9LU67 (Q9LU67) Phosphatidylserine decarb... 28 5.3
TC206399 similar to GB|BAB09797.1|9759182|AB013388 ATP-dependent... 28 5.3
CK768403 28 5.3
TC229698 similar to UP|Q9VKD8 (Q9VKD8) CG6686-PB (RE11167p), par... 28 5.3
BE800817 similar to SP|P00865|RBS1 Ribulose bisphosphate carboxy... 28 6.9
TC217405 similar to UP|Q9FLL9 (Q9FLL9) Gb|AAB80666.1, partial (20%) 28 6.9
>AW703863
Length = 408
Score = 256 bits (654), Expect = 1e-68
Identities = 121/135 (89%), Positives = 125/135 (91%)
Frame = +1
Query: 181 SFHFPIRSDPFVHRAIYSIADISVTPLTEGQIVNFERTYCQPLSDMNLPDSEGKNLHQYL 240
SFHFPIR D FV AIYSIADIS TPLTEGQIV+FE+TYCQPLSDMN+PDSEGKNLHQYL
Sbjct: 4 SFHFPIRYDLFVRGAIYSIADISGTPLTEGQIVHFEKTYCQPLSDMNIPDSEGKNLHQYL 183
Query: 241 KFNSNMGLIHIPCSEKYLPSENRKKGIPCLKKCLEKGQMYRTFQRDSVFSTLSKWNLFQT 300
FNSNMGLIH PCSEKYLPSE RKK PCLKKCLEKGQMYRTFQ+DSVFSTLSKWNLFQT
Sbjct: 184 NFNSNMGLIHTPCSEKYLPSEKRKKRSPCLKKCLEKGQMYRTFQQDSVFSTLSKWNLFQT 363
Query: 301 YIPWFLTSTGYKYLN 315
YIPWFLTSTGYKYLN
Sbjct: 364 YIPWFLTSTGYKYLN 408
>TC228876 similar to UP|YC2A_ATRBE (Q8S8V2) Protein ycf2, partial (5%)
Length = 610
Score = 113 bits (283), Expect = 1e-25
Identities = 60/78 (76%), Positives = 60/78 (76%), Gaps = 7/78 (8%)
Frame = +1
Query: 312 KYLNFIFLDTFSDLLPILSSSQKFVSIFHDIMHRSDISWRILQKKWCLSQ-------WNL 364
KYLNFIFLDTFSDLLPILSSSQKFVSIFHDIMH SDI WRI Q CL Q WNL
Sbjct: 1 KYLNFIFLDTFSDLLPILSSSQKFVSIFHDIMHGSDILWRIRQIPLCLPQWNLISEIWNL 180
Query: 365 ISEISSKCFHNLLLSEEI 382
ISEI C HNLLLSEEI
Sbjct: 181 ISEIPGYCLHNLLLSEEI 234
>CF923128
Length = 119
Score = 62.8 bits (151), Expect = 3e-10
Identities = 30/36 (83%), Positives = 32/36 (88%)
Frame = -1
Query: 142 KKHAFLERDTISPIESRVFNILILNDFPQSGDEGYN 177
KK FL+RDTISPIES+V NILI NDFPQSGDEGYN
Sbjct: 110 KKTCFLKRDTISPIESQVSNILIPNDFPQSGDEGYN 3
>CF922614
Length = 752
Score = 42.0 bits (97), Expect = 5e-04
Identities = 23/41 (56%), Positives = 29/41 (70%)
Frame = -2
Query: 144 HAFLERDTISPIESRVFNILILNDFPQSGDEGYNLYKSFHF 184
H FL+++ ISPI+S V LILNDF QS ++ YN YKS F
Sbjct: 739 HVFLKKNIISPIKS*V--TLILNDFSQSDNQRYN*YKSHTF 623
>TC219314 weakly similar to UP|Q7XUE7 (Q7XUE7) OJ991113_30.24 protein,
partial (5%)
Length = 898
Score = 33.1 bits (74), Expect(2) = 0.002
Identities = 14/23 (60%), Positives = 16/23 (68%)
Frame = -1
Query: 46 RKFFDNTDSYFSMISHDEDNWLN 68
R+ F D YF MISHD +NWLN
Sbjct: 739 REIF**VDPYFLMISHDPNNWLN 671
Score = 26.2 bits (56), Expect(2) = 0.002
Identities = 11/15 (73%), Positives = 12/15 (79%)
Frame = -3
Query: 69 PVKPFHRSSLISSFY 83
PVK FHRSSLI + Y
Sbjct: 671 PVKSFHRSSLILNIY 627
>BU548830
Length = 712
Score = 31.6 bits (70), Expect(2) = 0.004
Identities = 12/16 (75%), Positives = 13/16 (81%)
Frame = +1
Query: 53 DSYFSMISHDEDNWLN 68
D YF MISHD +NWLN
Sbjct: 19 DPYFLMISHDPNNWLN 66
Score = 26.2 bits (56), Expect(2) = 0.004
Identities = 11/15 (73%), Positives = 12/15 (79%)
Frame = +3
Query: 69 PVKPFHRSSLISSFY 83
PVK FHRSSLI + Y
Sbjct: 66 PVKSFHRSSLILNIY 110
>TC205093
Length = 1144
Score = 30.8 bits (68), Expect = 1.1
Identities = 12/44 (27%), Positives = 26/44 (58%)
Frame = +2
Query: 111 KACINNYDFTYGQFLNILFIRNKRFSLCGGKKKHAFLERDTISP 154
K CI+ Y F+ + L+ ++R++ CG + L+++T++P
Sbjct: 596 KNCISGYSFSPSKCLSQSWVRSRTTHCCGSICRKGVLQKETLAP 727
>TC223381
Length = 598
Score = 30.8 bits (68), Expect = 1.1
Identities = 18/69 (26%), Positives = 27/69 (39%)
Frame = -1
Query: 311 YKYLNFIFLDTFSDLLPILSSSQKFVSIFHDIMHRSDISWRILQKKWCLSQWNLISEISS 370
Y+ + FI F DLL S++KF S + +WC +N I
Sbjct: 232 YRRIVFILFPKFFDLLSSHPSTKKFTHFVQSFQLHSALFSLGHTIRWCYISYNSFCNIMH 53
Query: 371 KCFHNLLLS 379
+C+ N S
Sbjct: 52 ECYFNTSFS 26
>TC232786 similar to UP|Q947G7 (Q947G7) Thermal hysteresis protein STHP-64,
partial (10%)
Length = 430
Score = 29.6 bits (65), Expect = 2.4
Identities = 14/32 (43%), Positives = 19/32 (58%)
Frame = +3
Query: 320 DTFSDLLPILSSSQKFVSIFHDIMHRSDISWR 351
DT S LP L SSQ+F SI + + + +WR
Sbjct: 45 DTLSHELPRLQSSQEFPSIIREKVSKDGYNWR 140
>TC233718 similar to UP|Q9LTK1 (Q9LTK1) Arabidopsis thaliana genomic DNA,
chromosome 5, BAC clone:F17P19, partial (10%)
Length = 260
Score = 29.6 bits (65), Expect = 2.4
Identities = 19/53 (35%), Positives = 28/53 (51%), Gaps = 2/53 (3%)
Frame = +3
Query: 66 WLNPVKPFHRSSLISS-FYKANRLRF-LNNRYHFCFYCNKRLPFYVEKACINN 116
W+ +KPF R++LIS+ A R + L N + Y R FY+ K C+ N
Sbjct: 18 WVRALKPFDRATLISNEIISAVRSCYALCNE*KWSLYLLPRKLFYLHKFCMKN 176
>BG352463 similar to GP|11245494|gb| SPATULA {Arabidopsis thaliana}, partial
(19%)
Length = 471
Score = 28.9 bits (63), Expect = 4.0
Identities = 17/46 (36%), Positives = 23/46 (49%)
Frame = -1
Query: 319 LDTFSDLLPILSSSQKFVSIFHDIMHRSDISWRILQKKWCLSQWNL 364
LD S+ L+S++ SI + +I W L K CLS WNL
Sbjct: 270 LDASSNHFSSLTSAESITSIALEAGAALNILWPHLALKLCLSCWNL 133
>BI970256
Length = 703
Score = 28.9 bits (63), Expect = 4.0
Identities = 11/33 (33%), Positives = 16/33 (48%)
Frame = +2
Query: 71 KPFHRSSLISSFYKANRLRFLNNRYHFCFYCNK 103
KPF +++ L N+Y+ CF CNK
Sbjct: 296 KPFPSQNIVPGEQNLQELGIFQNQYY*CFLCNK 394
>CO980586
Length = 798
Score = 28.9 bits (63), Expect = 4.0
Identities = 11/36 (30%), Positives = 19/36 (52%)
Frame = +3
Query: 73 FHRSSLISSFYKANRLRFLNNRYHFCFYCNKRLPFY 108
+H+S +IS +Y ++ + YH YC K F+
Sbjct: 633 YHQSFIISYYYNDKKILIILK*YHMILYCYKIKSFF 740
>TC218379 similar to UP|Q9LU67 (Q9LU67) Phosphatidylserine decarboxylase,
partial (38%)
Length = 888
Score = 28.5 bits (62), Expect = 5.3
Identities = 18/65 (27%), Positives = 30/65 (45%)
Frame = -1
Query: 312 KYLNFIFLDTFSDLLPILSSSQKFVSIFHDIMHRSDISWRILQKKWCLSQWNLISEISSK 371
++ +F+++ T S +SS QK RS + WR L K Q N + + +
Sbjct: 765 QFQHFLYIKTISSWRHPISSLQK---------PRSPVIWRYLLKGLHQLQLNSLQKRKLQ 613
Query: 372 CFHNL 376
CFH +
Sbjct: 612 CFHRM 598
>TC206399 similar to GB|BAB09797.1|9759182|AB013388 ATP-dependent Clp
protease regulatory subunit CLPX {Arabidopsis thaliana;}
, partial (41%)
Length = 1207
Score = 28.5 bits (62), Expect = 5.3
Identities = 16/61 (26%), Positives = 26/61 (42%)
Frame = -1
Query: 56 FSMISHDEDNWLNPVKPFHRSSLISSFYKANRLRFLNNRYHFCFYCNKRLPFYVEKACIN 115
+S SH NW + SSL + F+K N + FC + N P+ + + +
Sbjct: 349 YSTSSHIGTNWCSKTNRRILSSLRNGFFKINNCSTTYKKNIFCVHLNVIPPWMLPSSFLR 170
Query: 116 N 116
N
Sbjct: 169 N 167
>CK768403
Length = 772
Score = 28.5 bits (62), Expect = 5.3
Identities = 17/45 (37%), Positives = 25/45 (54%), Gaps = 2/45 (4%)
Frame = -1
Query: 315 NFIFLDTFSDLLPILSSSQKFV--SIFHDIMHRSDISWRILQKKW 357
+F+F +FS P SSS F I H ++H + I +R Q+KW
Sbjct: 565 SFVFCFSFS---PSFSSSSCFPLPQIVHSVLH*NPIKFRNKQRKW 440
>TC229698 similar to UP|Q9VKD8 (Q9VKD8) CG6686-PB (RE11167p), partial (3%)
Length = 879
Score = 28.5 bits (62), Expect = 5.3
Identities = 12/26 (46%), Positives = 16/26 (61%)
Frame = +3
Query: 303 PWFLTSTGYKYLNFIFLDTFSDLLPI 328
P+F TSTG +L+ IF F +PI
Sbjct: 336 PYFFTSTGKPFLSIIFTFNFQSPIPI 413
>BE800817 similar to SP|P00865|RBS1 Ribulose bisphosphate carboxylase small
chain 1 chloroplast precursor (EC 4.1.1.39), partial
(85%)
Length = 493
Score = 28.1 bits (61), Expect = 6.9
Identities = 15/43 (34%), Positives = 21/43 (47%)
Frame = +2
Query: 263 RKKGIPCLKKCLEKGQMYRTFQRDSVFSTLSKWNLFQTYIPWF 305
RK IPCL+ L+ G MYR R + W ++ +P F
Sbjct: 266 RKGWIPCLESDLDHGFMYREHNRSPSYYDGRYWTMWN--LPMF 388
>TC217405 similar to UP|Q9FLL9 (Q9FLL9) Gb|AAB80666.1, partial (20%)
Length = 1652
Score = 28.1 bits (61), Expect = 6.9
Identities = 14/45 (31%), Positives = 25/45 (55%)
Frame = +2
Query: 296 NLFQTYIPWFLTSTGYKYLNFIFLDTFSDLLPILSSSQKFVSIFH 340
+L ++PWFL ++NF+ DLL I +S ++V++ H
Sbjct: 929 HLLMLHLPWFLHPG*VSFMNFL------DLLVINLASHEWVTLLH 1045
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.141 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,713,964
Number of Sequences: 63676
Number of extensions: 369014
Number of successful extensions: 2717
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 2697
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2714
length of query: 382
length of database: 12,639,632
effective HSP length: 99
effective length of query: 283
effective length of database: 6,335,708
effective search space: 1793005364
effective search space used: 1793005364
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0269b.10


