

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0269a.2
(447 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
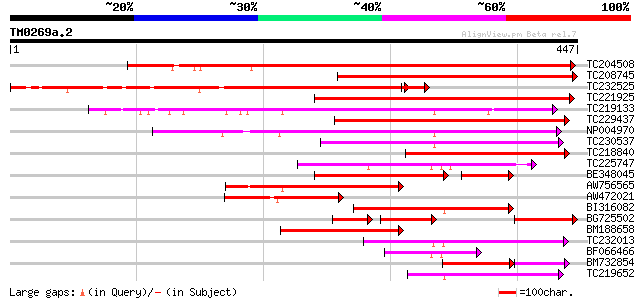
Score E
Sequences producing significant alignments: (bits) Value
TC204508 homologue to UP|Q39855 (Q39855) Cyclin, partial (98%) 555 e-158
TC208745 homologue to UP|CG22_SOYBN (P25012) G2/mitotic-specific... 323 7e-89
TC232525 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type,... 305 9e-87
TC221925 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type,... 254 6e-68
TC219133 UP|Q39879 (Q39879) Mitotic cyclin a2-type, complete 181 5e-46
TC229437 similar to UP|Q40670 (Q40670) Cyclin (Fragment), partia... 171 8e-43
NP004970 mitotic cyclin a1-type 147 1e-35
TC230537 mitotic cyclin a2-type 129 3e-30
TC218840 similar to UP|CG1B_MEDVA (P46277) G2/mitotic-specific c... 113 2e-25
TC225747 homologue to UP|Q6LBE8 (Q6LBE8) Histone H3.2 protein, c... 110 1e-24
BE348045 similar to PIR|T07672|T07 cyclin a2-type mitosis-speci... 90 2e-24
AW756565 similar to PIR|T07675|T07 cyclin a2-type mitosis-speci... 104 9e-23
AW472021 similar to PIR|S56679|S56 mitosis-specific cyclin CycII... 100 1e-21
BI316082 similar to PIR|T07672|T07 cyclin a2-type mitosis-speci... 99 5e-21
BG725502 similar to GP|1835260|emb| mitotic cyclin {Sesbania ros... 75 1e-20
BM188658 similar to PIR|T07669|T07 cyclin a1-type mitosis-speci... 94 1e-19
TC232013 similar to UP|Q9S957 (Q9S957) Cyclin A homolog, partial... 83 3e-16
BF066466 weakly similar to GP|22830757|dbj B1 type cyclin {Daucu... 77 2e-14
BM732854 similar to PIR|S56679|S56 mitosis-specific cyclin CycII... 60 5e-14
TC219652 similar to UP|Q39878 (Q39878) Mitotic cyclin a2-type, p... 68 1e-11
>TC204508 homologue to UP|Q39855 (Q39855) Cyclin, partial (98%)
Length = 1283
Score = 555 bits (1430), Expect = e-158
Identities = 287/366 (78%), Positives = 318/366 (86%), Gaps = 13/366 (3%)
Frame = +2
Query: 94 VAAPGVAVAKREAPKPVTKRVTGKPKPVVEVIEI--SPDEQIKKEKSVQKKKE---DSKK 148
VA GVAVAKR APKPV+K+V KPKP +V +I SPD KK KKKE + KK
Sbjct: 5 VANEGVAVAKRAAPKPVSKKVIVKPKPSEKVTDIDASPD---KKRVLKDKKKEGDANPKK 175
Query: 149 K---TLTSVLTARSKAACGLTNKPKEEIVDIDASDVDNELAAVE-----YIEDIYKFYKM 200
K TLTSVLTARSKAACG+TNKPKE+I+DIDASDVDNELAAVE YI+DIYKFYK+
Sbjct: 176 KSQHTLTSVLTARSKAACGITNKPKEQIIDIDASDVDNELAAVELAAVEYIDDIYKFYKL 355
Query: 201 VENESRPHCYMASQPEINEKMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVP 260
VENESRPH Y+ SQPEINE+MRAILVDWLIDVHTKFELSLETLYLTINI+DRFLAVKTVP
Sbjct: 356 VENESRPHDYIGSQPEINERMRAILVDWLIDVHTKFELSLETLYLTINIIDRFLAVKTVP 535
Query: 261 RRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVP 320
RRELQLVGIS+MLMA+KYEEIWPPEVNDFVCLSDRAY+HEQIL MEK IL +LEWTLTVP
Sbjct: 536 RRELQLVGISAMLMASKYEEIWPPEVNDFVCLSDRAYTHEQILAMEKTILNKLEWTLTVP 715
Query: 321 TPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKSP 380
TPFVFL RFIKA+VPD+ + NMAHF+SELGMM+Y TLMYCPSM+AASAV+AARCTLNK+P
Sbjct: 716 TPFVFLVRFIKAAVPDQELENMAHFMSELGMMNYATLMYCPSMVAASAVFAARCTLNKAP 895
Query: 381 AWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
WNETLKLHT YS+EQLMDCARLLV FH T+GNGKL+VV+RKYSDP++GAVAVLPPAK L
Sbjct: 896 LWNETLKLHTGYSQEQLMDCARLLVGFHSTLGNGKLRVVYRKYSDPQKGAVAVLPPAKLL 1075
Query: 441 MPQEAS 446
AS
Sbjct: 1076SEGSAS 1093
>TC208745 homologue to UP|CG22_SOYBN (P25012) G2/mitotic-specific cyclin
S13-7 (B-like cyclin) (Fragment), partial (74%)
Length = 879
Score = 323 bits (829), Expect = 7e-89
Identities = 153/189 (80%), Positives = 175/189 (91%)
Frame = +1
Query: 259 VPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLT 318
VPRRELQLVGIS+MLMA+KYEEIWPPEVNDFVCLSDRAY+HEQIL MEK IL +LEWTLT
Sbjct: 1 VPRRELQLVGISAMLMASKYEEIWPPEVNDFVCLSDRAYTHEQILAMEKTILNKLEWTLT 180
Query: 319 VPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNK 378
VPTPFVFL RFIKA+VPD+ + NMAHF+SELGMM+Y TLMYCPSM+AASAV+AARCTLNK
Sbjct: 181 VPTPFVFLVRFIKAAVPDQELENMAHFMSELGMMNYATLMYCPSMVAASAVFAARCTLNK 360
Query: 379 SPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAK 438
+P WNETLKLHT YS+EQLMDCARLLV FH T+ NGKL+VV+RKYSDP++GAVAVLPPAK
Sbjct: 361 APLWNETLKLHTGYSQEQLMDCARLLVGFHSTLENGKLRVVYRKYSDPQKGAVAVLPPAK 540
Query: 439 NLMPQEASS 447
L+P+ ++S
Sbjct: 541 FLLPEGSAS 567
>TC232525 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type, partial
(73%)
Length = 1242
Score = 305 bits (782), Expect(2) = 9e-87
Identities = 182/322 (56%), Positives = 220/322 (67%), Gaps = 7/322 (2%)
Frame = +1
Query: 1 MASRPVVPQQPRGDAVVGGGKQQPKKNGAAAGRNRRALGDIGNL--DPVRGIEVKPNRPI 58
MASR QQP VGG +Q KN GRNRR L DIGNL G + ++P+
Sbjct: 268 MASRLEQQQQPTN---VGGENKQ--KNMGGEGRNRRVLQDIGNLVGKQGHGNGINVSKPV 432
Query: 59 TRSFCAQLLANAQAAVAAENNKKQACPNVAGPPAPVAAPGVAVAKREAPKPVTKRVTGKP 118
TR+F AQLLANAQAA E NKK + G A VA GV V + V KP
Sbjct: 433 TRNFRAQLLANAQAAT--EKNKKSSTEVNNG--AVVATDGVGVGNFVPARKVG--AAKKP 594
Query: 119 KPVVEVIEISPDEQIKKEKSVQKKKEDSKK-----KTLTSVLTARSKAACGLTNKPKEEI 173
K EVI IS D++ +++++ + KKE K K +SVL+ARSKAACGL P++ +
Sbjct: 595 KEEPEVIVISSDDESEEKQAAKGKKEREKSARKNAKAFSSVLSARSKAACGL---PRDLV 765
Query: 174 VDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMASQPEINEKMRAILVDWLIDVH 233
++IDA+D+DNELAA EYI+DIYKFYK E E H YM SQP+IN KMR+ILVDWLI+VH
Sbjct: 766 MNIDATDMDNELAAAEYIDDIYKFYKETEEEGCVHDYMGSQPDINAKMRSILVDWLIEVH 945
Query: 234 TKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLS 293
KFEL ETLYLT+NIVDRFL+VK VPRRELQLVGISSML+A+KYEEIW PEVNDF C+S
Sbjct: 946 RKFELMPETLYLTLNIVDRFLSVKAVPRRELQLVGISSMLIASKYEEIWAPEVNDFECIS 1125
Query: 294 DRAYSHEQILVMEKIILGRLEW 315
D AY +Q+L+MEK IL +LEW
Sbjct: 1126DNAYVSQQVLMMEKKILRKLEW 1191
Score = 33.5 bits (75), Expect(2) = 9e-87
Identities = 15/22 (68%), Positives = 17/22 (77%)
Frame = +2
Query: 310 LGRLEWTLTVPTPFVFLTRFIK 331
LG L TLTVPTP+ FL R+IK
Sbjct: 1175 LGSLNGTLTVPTPYHFLVRYIK 1240
>TC221925 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type, partial
(46%)
Length = 802
Score = 254 bits (649), Expect = 6e-68
Identities = 126/207 (60%), Positives = 161/207 (76%), Gaps = 2/207 (0%)
Frame = +2
Query: 241 ETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHE 300
ETLYLT+ IVDRFL+VK VPRRELQLVGISSML+A+KYEEIW PEVNDF C+SD AY +
Sbjct: 5 ETLYLTLXIVDRFLSVKAVPRRELQLVGISSMLIASKYEEIWAPEVNDFECISDNAYVSQ 184
Query: 301 QILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVP-DEGVTNMAHFLSELGMMHYDT-LM 358
Q+L+ME IL +LEW LTVPTP+ FL R+IKAS P D+ + NM FL+ELG+MHY T ++
Sbjct: 185 QVLMMEXXILRKLEWYLTVPTPYHFLVRYIKASTPSDKEMENMVFFLAELGLMHYPTAIL 364
Query: 359 YCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKV 418
Y PS+IAA+AV+AARCTL +SP W TLK +T YSEEQL DCA+++V+ H KL+
Sbjct: 365 YRPSLIAAAAVFAARCTLGRSPFWTSTLKHYTGYSEEQLRDCAKIMVNLHAAAPGSKLRA 544
Query: 419 VFRKYSDPERGAVAVLPPAKNLMPQEA 445
V++K+ + + AVA+L PAK ++A
Sbjct: 545 VYKKFCNSDLSAVALLSPAKENCLEQA 625
>TC219133 UP|Q39879 (Q39879) Mitotic cyclin a2-type, complete
Length = 1804
Score = 181 bits (460), Expect = 5e-46
Identities = 134/410 (32%), Positives = 207/410 (49%), Gaps = 40/410 (9%)
Frame = +2
Query: 63 CAQLLANAQAAV--AAENNKKQACPNVAGPPAPVAAPGVAV--AKREAPK---PVTKRVT 115
CA A + V A NK+ NV P + V P K EAP P VT
Sbjct: 230 CAAKFAKTKKDVVPACSGNKRPTLANVK-PASAVIFPKANSFPVKNEAPPTPPPPVATVT 406
Query: 116 GKPKPVVEV-------IEISPDEQIKK---EKSVQKKKEDSKKKTLTSVLTARSKAACGL 165
P PVV+V + +S DE + KS + D+ + ++ + +
Sbjct: 407 -VPAPVVDVSPSKSDTMSVSTDESMSSCDSFKSPDIEYVDNSDVAAVDSIERKTFSHLNI 583
Query: 166 TNKP------KEEIVDIDASD----VDNELA----AVEYIEDIYKFYKMVENESRPHCYM 211
++ +E +V+++ D VDN A + DIYK + E + RP
Sbjct: 584 SDSTEGNICSREILVELEKGDKFVNVDNNYADPQLCATFACDIYKHLRASEAKKRPSTDF 763
Query: 212 AS--QPEINEKMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGI 269
Q EIN MRAIL+DWL++V ++ L +TLYLT+N +DR+L+ + R+ LQL+G+
Sbjct: 764 MEKIQKEINSSMRAILIDWLVEVAEEYRLVPDTLYLTVNYIDRYLSGNVMNRQRLQLLGV 943
Query: 270 SSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRF 329
+SM++A+KYEEI P+V +F ++D Y E++L ME +L L++ +T PT FL RF
Sbjct: 944 ASMMIASKYEEICAPQVEEFCYITDNTYFKEEVLQMESAVLNFLKFEMTAPTVKCFLRRF 1123
Query: 330 IKAS-----VPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTL--NKSPAW 382
++A+ VP + + ++++EL +M Y L Y PS++AASA++ A+ L +K P W
Sbjct: 1124VRAAQGVDEVPSLQLECLTNYIAELSLMEYSMLGYAPSLVAASAIFLAKFILFPSKKP-W 1300
Query: 383 NETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVA 432
N TL+ +T Y L C + L C N L + KYS + VA
Sbjct: 1301NSTLQHYTLYQPSDLCVCVKDLHRLCCNSPNSNLPAIREKYSQHKYKYVA 1450
>TC229437 similar to UP|Q40670 (Q40670) Cyclin (Fragment), partial (77%)
Length = 830
Score = 171 bits (432), Expect = 8e-43
Identities = 79/185 (42%), Positives = 134/185 (71%)
Frame = +1
Query: 257 KTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWT 316
+ V R++LQLVG+++ML+A KYEE+ P V DF+ ++D+AY+ ++L MEK+++ L++
Sbjct: 7 QAVIRKKLQLVGVTAMLIACKYEEVSVPTVEDFILITDKAYTRNEVLDMEKLMMNILQFK 186
Query: 317 LTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTL 376
L+VPTP++F+ RF+KA+ D+ + ++ FL EL ++ L + PS++AA+A+Y A+C+L
Sbjct: 187 LSVPTPYMFMRRFLKAAHSDKKLELLSFFLVELCLVECKMLKFSPSLLAAAAIYTAQCSL 366
Query: 377 NKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPP 436
+ W +T + +TDYSEE+L++C+RL+V+FH G+GKL V+RKY+ + G A + P
Sbjct: 367 YQFKQWTKTTEWYTDYSEEKLLECSRLMVTFHQKAGSGKLTGVYRKYNTWKYGCAAKIEP 546
Query: 437 AKNLM 441
A L+
Sbjct: 547 ALFLL 561
>NP004970 mitotic cyclin a1-type
Length = 1047
Score = 147 bits (370), Expect = 1e-35
Identities = 92/337 (27%), Positives = 170/337 (50%), Gaps = 14/337 (4%)
Frame = +1
Query: 113 RVTGKPKPVVEVIEISPDEQIKKEKSVQKKKEDSKKKTLTSVLTARS-KAACGLT---NK 168
R K K ++ + + K+++ V + + + ++ + R K C NK
Sbjct: 10 RAAAKRKANAAIVVVVEKQHPKRQRVVLGELPNLQNLIVSKIQNPRKEKLQCRKNPNANK 189
Query: 169 PKEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYM----ASQPEINEKMRAI 224
P + + +D Y+ DI+++ + +E + + + Q + MR I
Sbjct: 190 PSPTNNTLSSPQLDGS-----YVSDIHEYLREMEMQKKRRPMVNYIEKFQKIVTPTMRGI 354
Query: 225 LVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPP 284
LVDWL++V +++L +TL+L+++ +DRFL+V V + LQL+G+SSML+AAKYEE PP
Sbjct: 355 LVDWLVEVAEEYKLLSDTLHLSVSYIDRFLSVNPVTKSRLQLLGVSSMLIAAKYEETDPP 534
Query: 285 EVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKAS-----VPDEGV 339
V++F ++D Y +++ ME IL L++ + PT FL R+ + P+ +
Sbjct: 535 SVDEFCSITDNTYDKAEVVKMEADILKSLKFEMGNPTVSTFLRRYANVASDVQKTPNSQI 714
Query: 340 TNMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTL-NKSPAWNETLKLHTDYSEEQLM 398
++ ++ EL ++ YD L + PS++AAS ++ A+ + + W +L + Y +L
Sbjct: 715 EHLGSYIGELSLLDYDCLRFLPSIVAASVIFLAKFIIWPEVHPWTSSLCECSGYKPAELK 894
Query: 399 DCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLP 435
+C +L + + K V KY + VA LP
Sbjct: 895 ECVLILHDLYLSRKAASFKAVREKYKHQKFKCVANLP 1005
>TC230537 mitotic cyclin a2-type
Length = 993
Score = 129 bits (323), Expect = 3e-30
Identities = 69/197 (35%), Positives = 116/197 (58%), Gaps = 6/197 (3%)
Frame = +1
Query: 246 TINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVM 305
++N++DR+L+ + + +++LQL+G++ ML+A+KYEE+ P V +F ++D Y+ E++L M
Sbjct: 7 SVNLIDRYLSTRLIQKQKLQLLGVTCMLIASKYEEMCAPRVEEFCFITDNTYTKEEVLKM 186
Query: 306 EKIILGRLEWTLTVPTPFVFLTRFIKAS-----VPDEGVTNMAHFLSELGMMHYDTLMYC 360
E+ +L + + L+VPT FL RFI+A+ P + +A++L+EL ++ + +
Sbjct: 187 EREVLDLVHFQLSVPTIKTFLRRFIQAAQSSYKAPCVELEFLANYLAELALVECNFFQFL 366
Query: 361 PSMIAASAVYAARCTLNKSP-AWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVV 419
PS++AASAV+ A+ TLN+S WN TLK +T Y +L L L V
Sbjct: 367 PSLVAASAVFLAKWTLNESEHPWNPTLKHYTKYKASELKTVVLALQDLQLNTKGSSLNAV 546
Query: 420 FRKYSDPERGAVAVLPP 436
KY + VA L P
Sbjct: 547 PEKYKQQKFNCVANLSP 597
>TC218840 similar to UP|CG1B_MEDVA (P46277) G2/mitotic-specific cyclin 1
(B-like cyclin) (CycMs1), partial (32%)
Length = 922
Score = 113 bits (282), Expect = 2e-25
Identities = 53/129 (41%), Positives = 86/129 (66%)
Frame = +1
Query: 313 LEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFLSELGMMHYDTLMYCPSMIAASAVYAA 372
L++ ++VPT +VF+ RF+KA+ D + +A FL EL ++ Y+ L + PS++AA+AVY A
Sbjct: 10 LQFNMSVPTAYVFMKRFLKAAQADRKLELLAFFLVELTLVEYEMLKFPPSLLAAAAVYTA 189
Query: 373 RCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVA 432
+CT+ W++T + H++YSE+QL++C+ L+ +FH GNGKL V RKY + A
Sbjct: 190 QCTIYGFKQWSKTCEWHSNYSEDQLLECSTLMAAFHQKAGNGKLTGVHRKYCSSKFSYTA 369
Query: 433 VLPPAKNLM 441
PA+ L+
Sbjct: 370 KCEPARFLL 396
>TC225747 homologue to UP|Q6LBE8 (Q6LBE8) Histone H3.2 protein, complete
Length = 1486
Score = 110 bits (276), Expect = 1e-24
Identities = 84/242 (34%), Positives = 115/242 (46%), Gaps = 54/242 (22%)
Frame = -2
Query: 228 WLIDVHT-KFELSLETLY-LTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEI---- 281
WL ++ +++L E INI+DR+L V +ELQL+GISS+L +Y +
Sbjct: 1386 WLSEIRKYRYQLLCEIAQNFKINIIDRYLWSTIVL*KELQLLGISSLLSMKRYCLLR*LA 1207
Query: 282 -----------------WPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFV 324
P V DF+ +S+ AYS IL MEK IL RLEW TVPTP+V
Sbjct: 1206 LTFT**HTEHFILSS**LKP*VKDFITISNDAYSRNHILSMEKAILERLEWNFTVPTPYV 1027
Query: 325 FLTRFIK--ASVPDEGV-----------TNMAHFL-----------------SELGMMHY 354
FL RFI+ PD+ V ++F+ ++HY
Sbjct: 1026 FLVRFIRTFGLPPDQQVIK*TST*TLQLIKYSYFIL*IAFALCRKKK*HFS*QNFSLVHY 847
Query: 355 DTL-MYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLMDCARLLVSFHCTVGN 413
+T + PSM AA+AVYAARCTLN+ WNE LK Y+ QL + CT+
Sbjct: 846 ETANLSPPSMTAAAAVYAARCTLNRKHPWNEILKDMAGYTTAQLRE--------FCTLDR 691
Query: 414 GK 415
G+
Sbjct: 690 GR 685
>BE348045 similar to PIR|T07672|T07 cyclin a2-type mitosis-specific -
soybean, partial (34%)
Length = 491
Score = 89.7 bits (221), Expect(2) = 2e-24
Identities = 45/106 (42%), Positives = 69/106 (64%)
Frame = -1
Query: 241 ETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVNDFVCLSDRAYSHE 300
+TLYLT+N++DR L+ V ++ LQL+G++ ML+A+KYEEI P V +F ++D Y+
Sbjct: 488 DTLYLTVNLIDRSLSQSLVQKQRLQLLGVTCMLIASKYEEICAPRVEEFCFITDNTYTKA 309
Query: 301 QILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPDEGVTNMAHFL 346
++L ME +L L + L+VPT FL RFI AS + ++ FL
Sbjct: 308 EVLKMESEVLNLLHFQLSVPTTKTFLRRFILASQSSYKLLAVSTFL 171
Score = 41.2 bits (95), Expect(2) = 2e-24
Identities = 21/42 (50%), Positives = 29/42 (69%), Gaps = 1/42 (2%)
Frame = -2
Query: 357 LMYCPSMIAASAVYAARCTLNKSP-AWNETLKLHTDYSEEQL 397
L + PS+IAASAV AR TLN+S WN T++ +T+Y +L
Sbjct: 190 LQFLPSLIAASAVLLARWTLNQSEHPWNSTMEHYTNYKVSEL 65
>AW756565 similar to PIR|T07675|T07 cyclin a2-type mitosis-specific -
soybean, partial (28%)
Length = 472
Score = 104 bits (259), Expect = 9e-23
Identities = 57/142 (40%), Positives = 88/142 (61%), Gaps = 2/142 (1%)
Frame = +2
Query: 171 EEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHCYMAS--QPEINEKMRAILVDW 228
++IV+ID D +L A Y+ DIYK + + RP Q +IN MR ILVDW
Sbjct: 47 DKIVNIDNIYSDTQLCAT-YVCDIYKHLRES*EKKRPSTDFMDTIQKDINVSMRTILVDW 223
Query: 229 LIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSMLMAAKYEEIWPPEVND 288
L++V ++ L ETLYLT+N +DR L+ + R+ +QL+G+S M++A+KYEEI P V +
Sbjct: 224 LVEVAEEYRLVPETLYLTVNYLDRSLSWNAMNRQRIQLLGVSCMMIASKYEEICAPHVEE 403
Query: 289 FVCLSDRAYSHEQILVMEKIIL 310
+ ++D Y E++L ME ++
Sbjct: 404 YRYITDDTYLKEEVLQMESCVV 469
>AW472021 similar to PIR|S56679|S56 mitosis-specific cyclin CycIII - alfalfa,
partial (24%)
Length = 317
Score = 100 bits (249), Expect = 1e-21
Identities = 51/98 (52%), Positives = 70/98 (71%), Gaps = 4/98 (4%)
Frame = +2
Query: 170 KEEIVDIDASDVDNELAAVEYIEDIYKFYKMVENESRPHC----YMASQPEINEKMRAIL 225
+E ++DID D +N LA V+YIED+Y Y+ +E S C YMA Q +INE+MRAIL
Sbjct: 32 EETVLDIDTCDANNPLAVVDYIEDLYAHYRKMEGTS---CVSPDYMAQQFDINERMRAIL 202
Query: 226 VDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRE 263
+DWLI+ H K +L ETL LT+N++DRFLA +TV R++
Sbjct: 203 IDWLIEGHDKLDLLHETLVLTVNLIDRFLAKQTVVRKK 316
>BI316082 similar to PIR|T07672|T07 cyclin a2-type mitosis-specific -
soybean, partial (31%)
Length = 456
Score = 98.6 bits (244), Expect = 5e-21
Identities = 56/132 (42%), Positives = 82/132 (61%), Gaps = 6/132 (4%)
Frame = +3
Query: 272 MLMAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIK 331
ML+A+KYEEI P V +F ++D Y+ ++L ME +L L + L+VPT FL RFI
Sbjct: 6 MLIASKYEEICAPRVEEFCFITDNTYTKAEVLKMESEVLNLLHFQLSVPTTKTFLRRFIL 185
Query: 332 ASVPDEGVTN-----MAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKSP-AWNET 385
AS V+ +A++L+EL ++ Y L + PS+IAASAV AR TLN+S WN T
Sbjct: 186 ASESSYKVSYVELEFLANYLAELTLVEYSFLQFLPSLIAASAVLLARWTLNQSEHPWNST 365
Query: 386 LKLHTDYSEEQL 397
++ +T+Y +L
Sbjct: 366 MEHYTNYKVSEL 401
>BG725502 similar to GP|1835260|emb| mitotic cyclin {Sesbania rostrata},
partial (20%)
Length = 423
Score = 75.1 bits (183), Expect = 6e-14
Identities = 34/49 (69%), Positives = 42/49 (85%)
Frame = +1
Query: 399 DCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNLMPQEASS 447
DCARL VSFH TVGN + KVV+ KYSDPE+G VA+LPPAKNL+P+ ++S
Sbjct: 229 DCARLFVSFHSTVGNREEKVVYLKYSDPEKGVVAMLPPAKNLLPEGSNS 375
Score = 72.0 bits (175), Expect(2) = 1e-20
Identities = 34/44 (77%), Positives = 39/44 (88%)
Frame = +3
Query: 293 SDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKASVPD 336
S+RAY+ EQILVM+KIIL +LEWTLT+PTP VFL FIKASVPD
Sbjct: 90 SNRAYTGEQILVMDKIILAKLEWTLTMPTPLVFLLHFIKASVPD 221
Score = 46.2 bits (108), Expect(2) = 1e-20
Identities = 21/32 (65%), Positives = 27/32 (83%)
Frame = +1
Query: 255 AVKTVPRRELQLVGISSMLMAAKYEEIWPPEV 286
AVK VP+ LQLV IS++LMA KYEEI+PP++
Sbjct: 1 AVKNVPKS*LQLVDISALLMATKYEEIYPPQI 96
>BM188658 similar to PIR|T07669|T07 cyclin a1-type mitosis-specific -
soybean, partial (32%)
Length = 438
Score = 94.0 bits (232), Expect = 1e-19
Identities = 43/97 (44%), Positives = 69/97 (70%)
Frame = +3
Query: 214 QPEINEKMRAILVDWLIDVHTKFELSLETLYLTINIVDRFLAVKTVPRRELQLVGISSML 273
Q ++ MR ILVDWL++V +++L +TL+L+++ +DRFL+V V + LQL+G+SSML
Sbjct: 144 QKQVTTTMRTILVDWLVEVAEEYKLLSDTLHLSVSYIDRFLSVNPVSKSRLQLLGVSSML 323
Query: 274 MAAKYEEIWPPEVNDFVCLSDRAYSHEQILVMEKIIL 310
+AAKYEE+ PP V+ F ++D Y +++ ME +L
Sbjct: 324 IAAKYEEVDPPRVDPFCNITDNTYHKAEVVKMEADML 434
>TC232013 similar to UP|Q9S957 (Q9S957) Cyclin A homolog, partial (38%)
Length = 814
Score = 82.8 bits (203), Expect = 3e-16
Identities = 57/167 (34%), Positives = 84/167 (50%), Gaps = 6/167 (3%)
Frame = +1
Query: 280 EIWPPEVNDFVCLSDRAYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIKA---SVPD 336
EI P + DF ++D Y+ ++L ME+ +L E+ L PT F+ RF++A S D
Sbjct: 1 EINAPRIEDFCFITDNTYTKAEVLKMERQVLKSSEYQLFAPTIQTFVRRFLRAAQASYKD 180
Query: 337 EGVT--NMAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKS-PAWNETLKLHTDYS 393
+ + +A++L+EL +M Y L + PS+IAASAV+ AR TL++S WN TL+ + Y
Sbjct: 181 QSLELEYLANYLAELTLMDYGFLNFLPSIIAASAVFLARWTLDQSNHPWNPTLQHYACYK 360
Query: 394 EEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNL 440
L L L V KY + VA L K L
Sbjct: 361 ASDLKTTVLALQDLQLNTDGCPLTAVRTKYRQDKFKCVAALSSPKLL 501
>BF066466 weakly similar to GP|22830757|dbj B1 type cyclin {Daucus carota},
partial (15%)
Length = 333
Score = 77.0 bits (188), Expect = 2e-14
Identities = 51/109 (46%), Positives = 59/109 (53%), Gaps = 32/109 (29%)
Frame = -1
Query: 296 AYSHEQILVMEKIILGRLEWTLTVPTPFVFLTRFIK--ASVPDEGV-------------- 339
AYS QIL MEK IL RLEW LTVPTP+VFL RF + A PD+ V
Sbjct: 330 AYSRIQILSMEKAILERLEWKLTVPTPYVFLVRFTRTFALSPDQQVIVIYLNSNTKNYQI 151
Query: 340 ---------------TNMAHFLSELGMMHYDTL-MYCPSMIAASAVYAA 372
NMA FL+ELG +HY T ++ PSM AA+AVYAA
Sbjct: 150 QLTCFACHLIHIVQMKNMAFFLAELGRVHYGTANLFLPSMTAAAAVYAA 4
>BM732854 similar to PIR|S56679|S56 mitosis-specific cyclin CycIII - alfalfa,
partial (25%)
Length = 421
Score = 59.7 bits (143), Expect(2) = 5e-14
Identities = 25/57 (43%), Positives = 42/57 (72%)
Frame = +3
Query: 342 MAHFLSELGMMHYDTLMYCPSMIAASAVYAARCTLNKSPAWNETLKLHTDYSEEQLM 398
+A FL EL ++ Y+ L + PS++AA+AVY A+CT+ W++T + H++YSE+QL+
Sbjct: 21 LAFFLVELSLVEYEMLKFPPSLLAAAAVYTAQCTIYGFKQWSKTCEWHSNYSEDQLL 191
Score = 35.8 bits (81), Expect(2) = 5e-14
Identities = 17/43 (39%), Positives = 24/43 (55%)
Frame = +1
Query: 399 DCARLLVSFHCTVGNGKLKVVFRKYSDPERGAVAVLPPAKNLM 441
+C+ L+ +FH GNGKL V RKY + A PA+ L+
Sbjct: 280 ECSTLMAAFHQKAGNGKLTGVHRKYCSSKFSYTAKCEPARFLL 408
>TC219652 similar to UP|Q39878 (Q39878) Mitotic cyclin a2-type, partial (30%)
Length = 670
Score = 67.8 bits (164), Expect = 1e-11
Identities = 45/129 (34%), Positives = 69/129 (52%), Gaps = 6/129 (4%)
Frame = +1
Query: 314 EWTLTVPTPFVFLTRFIKASVPDEGVTN-----MAHFLSELGMMHYDTLMYCPSMIAASA 368
++ L+VPT FL RFI AS V+ +A++L+EL ++ Y L + PS+IAASA
Sbjct: 46 DFQLSVPTTKTFLRRFILASQSSYKVSYVELEFLANYLAELTLVEYSFLQFLPSLIAASA 225
Query: 369 VYAARCTLNKSP-AWNETLKLHTDYSEEQLMDCARLLVSFHCTVGNGKLKVVFRKYSDPE 427
V AR TLN+S WN T++ +T+Y +L L + + + KY+ +
Sbjct: 226 VLLARWTLNQSEHPWNSTMEHYTNYKVSELKTAVLALADLLHDMKGCSVHSIREKYTQQK 405
Query: 428 RGAVAVLPP 436
+VA L P
Sbjct: 406 FRSVANLSP 432
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,406,736
Number of Sequences: 63676
Number of extensions: 255922
Number of successful extensions: 4162
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 1894
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2791
length of query: 447
length of database: 12,639,632
effective HSP length: 100
effective length of query: 347
effective length of database: 6,272,032
effective search space: 2176395104
effective search space used: 2176395104
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0269a.2


