

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
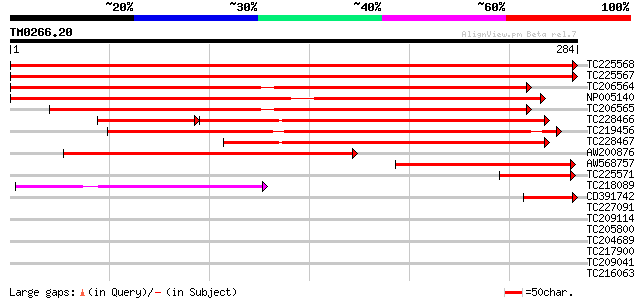
Query= TM0266.20
(284 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225568 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp pro... 535 e-153
TC225567 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp pro... 535 e-152
TC206564 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein... 288 3e-78
NP005140 heat shock protein 268 2e-72
TC206565 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-l... 262 1e-70
TC228466 similar to UP|ERD1_ARATH (P42762) ERD1 protein, chlorop... 179 3e-66
TC219456 homologue to UP|Q9LLI0 (Q9LLI0) ClpB, partial (24%) 223 8e-59
TC228467 weakly similar to UP|ERD1_ARATH (P42762) ERD1 protein, ... 162 1e-40
AW200876 weakly similar to GP|4105131|gb| ClpC protease {Spinaci... 146 1e-35
AW568757 similar to SP|P35100|CLPA_ ATP-dependent clp protease A... 140 9e-34
TC225571 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp prote... 60 9e-10
TC218089 52 2e-07
CD391742 homologue to GP|15912291|gb| AT4g33680/T16L1_170 {Arabi... 51 7e-07
TC227091 similar to UP|Q7Q6L1 (Q7Q6L1) AgCP5440 (Fragment), part... 33 0.15
TC209114 similar to GB|AAP21288.1|30102740|BT006480 At1g52360 {A... 33 0.19
TC205800 similar to GB|AAP21288.1|30102740|BT006480 At1g52360 {A... 31 0.73
TC204689 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type A... 30 0.95
TC217900 UP|O48924 (O48924) CYP83D1p (Fragment), complete 30 1.6
TC209041 weakly similar to UP|Q9V780 (Q9V780) Lap1 protein, part... 29 2.8
TC216063 UP|C822_SOYBN (O81972) Cytochrome P450 82A2 (P450 CP4)... 28 6.2
>TC225568 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (36%)
Length = 1287
Score = 535 bits (1378), Expect = e-153
Identities = 273/284 (96%), Positives = 279/284 (98%)
Frame = +2
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SFIFSGPTGVGKSELAK LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG
Sbjct: 158 SFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 337
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN
Sbjct: 338 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 517
Query: 121 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 180
VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK
Sbjct: 518 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 697
Query: 181 LEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDS 240
LEVKEIADIMLKEVFDRLK KEIDLSVTERFR+RVV+EGY+PSYGARPLRRAIMRLLEDS
Sbjct: 698 LEVKEIADIMLKEVFDRLKAKEIDLSVTERFRERVVDEGYNPSYGARPLRRAIMRLLEDS 877
Query: 241 MAEKMLAGEIKEGDSVIIDADSDGKVIVLNGSSGAPESLPEALP 284
MAEKMLA EIKEGDSVI+DADSDG VIVLNGSSGAP+SL EALP
Sbjct: 878 MAEKMLAREIKEGDSVIVDADSDGNVIVLNGSSGAPDSLEEALP 1009
>TC225567 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor ,
partial (61%)
Length = 2077
Score = 535 bits (1377), Expect = e-152
Identities = 272/284 (95%), Positives = 277/284 (96%)
Frame = +3
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SFIFSGPTGVGKSELAK LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG
Sbjct: 858 SFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 1037
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN
Sbjct: 1038 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 1217
Query: 121 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 180
VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK
Sbjct: 1218 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 1397
Query: 181 LEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDS 240
LEVKEIADIMLKEVFDRLK K+I+L VTERFRDRVVEEGY+PSYGARPLRRAIMRLLEDS
Sbjct: 1398 LEVKEIADIMLKEVFDRLKVKDIELQVTERFRDRVVEEGYNPSYGARPLRRAIMRLLEDS 1577
Query: 241 MAEKMLAGEIKEGDSVIIDADSDGKVIVLNGSSGAPESLPEALP 284
MAEKMLA EIKEGDSVI+D DSDG VIVLNGSSGAPESLPE LP
Sbjct: 1578 MAEKMLAREIKEGDSVIVDVDSDGNVIVLNGSSGAPESLPETLP 1709
>TC206564 homologue to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like,
partial (40%)
Length = 1497
Score = 288 bits (736), Expect = 3e-78
Identities = 141/261 (54%), Positives = 192/261 (73%)
Frame = +2
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SF+F GPTGVGK+ELAK LA+Y F +EEA++R+DMSE+ME+H VS+LIG+PPGYVGY EG
Sbjct: 365 SFMFMGPTGVGKTELAKALAAYLFNTEEALVRIDMSEYMEKHAVSRLIGAPPGYVGYEEG 544
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTE VRRRPY V+LFDEIEKAH DVFN+ LQIL+DGR+TDS+GRTV F NT++IMTSN
Sbjct: 545 GQLTEIVRRRPYAVILFDEIEKAHADVFNVFLQILDDGRVTDSQGRTVSFTNTVIIMTSN 724
Query: 121 VGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTK 180
VGS I + D D K+ +Y IK V + + FRPEF+NR+DE IVF+ L +
Sbjct: 725 VGSQYI------LNTDDDTTPKELAYETIKQRVMDAARSIFRPEFMNRVDEYIVFQPLDR 886
Query: 181 LEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDS 240
++ I + L+ V R+ +++ + VT+ + GYDP+YGARP++R I + +E+
Sbjct: 887 EQISSIVRLQLERVQKRIADRKMKIQVTDAAVQLLGSLGYDPNYGARPVKRVIQQNVENE 1066
Query: 241 MAEKMLAGEIKEGDSVIIDAD 261
+A+ +L GE KE D+++ID +
Sbjct: 1067LAKGILRGEFKEEDAILIDTE 1129
>NP005140 heat shock protein
Length = 2736
Score = 268 bits (685), Expect = 2e-72
Identities = 136/269 (50%), Positives = 191/269 (70%), Gaps = 1/269 (0%)
Frame = +1
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
SF+F GPTGVGK+ELAK LA F +E ++R+DMSE+ME+H+VS+LIG+PPGYVG+ EG
Sbjct: 1801 SFLFLGPTGVGKTELAKALAEQLFDNENQLVRIDMSEYMEQHSVSRLIGAPPGYVGHEEG 1980
Query: 61 GQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 120
GQLTEAVRRRPY+VVLFDE+EKAH VFN +LQ+L+DGRLTD +GRTVDF+NT++IMTSN
Sbjct: 1981 GQLTEAVRRRPYSVVLFDEVEKAHTSVFNTLLQVLDDGRLTDGQGRTVDFRNTVIIMTSN 2160
Query: 121 VGSSVIEKG-GRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLT 179
+G+ + G K + D V +E+++ FRPE LNRLDE++VF L+
Sbjct: 2161 LGAEHLLSGLSGKCTMQVARDR-----------VMQEVRRQFRPELLNRLDEIVVFDPLS 2307
Query: 180 KLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLED 239
+++++A + +K+V RL K I L+VT+ D ++ E YDP YGARP+RR + + +
Sbjct: 2308 HDQLRKVARLQMKDVASRLAEKGIALAVTDAALDYILSESYDPVYGARPIRRWLEKKVVT 2487
Query: 240 SMAEKMLAGEIKEGDSVIIDADSDGKVIV 268
++ ++ EI E +V IDA +G +V
Sbjct: 2488 ELSRMLVREEIDENSTVYIDAGPNGGELV 2574
>TC206565 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like, partial
(25%)
Length = 1078
Score = 262 bits (669), Expect = 1e-70
Identities = 128/241 (53%), Positives = 176/241 (72%)
Frame = +2
Query: 21 SYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEI 80
SY F +EEA++R+DMSE+ME+HTVS+LIG+PPGYVGY EGGQLTE VRRRPY V+LFDEI
Sbjct: 2 SYLFNTEEALVRIDMSEYMEKHTVSRLIGAPPGYVGYEEGGQLTETVRRRPYAVILFDEI 181
Query: 81 EKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYD 140
EKAH DVFN+ LQIL+DGR+TDS+GRTV F NT++IMTSNVGS I + D D
Sbjct: 182 EKAHSDVFNVFLQILDDGRVTDSQGRTVSFTNTVIIMTSNVGSQYI------LNTDDDTV 343
Query: 141 EKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKT 200
K+S+Y IK V + + FRPEF+NR+DE IVF+ L + ++ I + L+ V R+
Sbjct: 344 PKESAYETIKQRVMDAARSIFRPEFMNRVDEYIVFQPLDRNQISSIVRLQLERVQKRIAD 523
Query: 201 KEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDA 260
+++ + VTE + GYDP+YGARP++R I + +E+ +A+ +L GE KE D++++D
Sbjct: 524 RKMKIQVTEAAIQLLGSLGYDPNYGARPVKRVIQQNVENELAKGILRGEFKEEDTILVDT 703
Query: 261 D 261
+
Sbjct: 704 E 706
>TC228466 similar to UP|ERD1_ARATH (P42762) ERD1 protein, chloroplast
precursor, partial (23%)
Length = 936
Score = 179 bits (454), Expect(2) = 3e-66
Identities = 90/176 (51%), Positives = 131/176 (74%), Gaps = 1/176 (0%)
Frame = +2
Query: 96 EDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRK-IGFDLDYDEKDSSYNRIKSLVT 154
+DG+LTDS+GR V FKN L++MTSNVGSS I KG IGF L D+K +SYN +KS+V
Sbjct: 155 QDGQLTDSQGRRVSFKNALVVMTSNVGSSAIAKGRHNSIGF-LIPDDKTTSYNGLKSMVI 331
Query: 155 EELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDR 214
EEL+ YFRPE LNR+DE++VF+ L K ++ +I D++L+++ R+ + + + V+E ++
Sbjct: 332 EELRSYFRPELLNRIDEVVVFQPLEKSQLLQILDLLLQDMKKRVLSLGVHVKVSEAVKNL 511
Query: 215 VVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLN 270
V ++GY+P+YGARPLRRAI L+ED ++E L GE K+GD+V+ID D++G V N
Sbjct: 512 VCQQGYNPTYGARPLRRAITSLIEDPLSEAFLYGECKQGDTVLIDLDANGNPFVTN 679
Score = 90.5 bits (223), Expect(2) = 3e-66
Identities = 40/51 (78%), Positives = 48/51 (93%)
Frame = +1
Query: 45 SKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQIL 95
SKLIGSPPGYVGY EGG LTEA+RR+P+T++L DEIEKAHPD+FN++LQIL
Sbjct: 1 SKLIGSPPGYVGYGEGGVLTEAIRRKPFTLLLLDEIEKAHPDIFNILLQIL 153
>TC219456 homologue to UP|Q9LLI0 (Q9LLI0) ClpB, partial (24%)
Length = 871
Score = 223 bits (568), Expect = 8e-59
Identities = 114/227 (50%), Positives = 161/227 (70%)
Frame = +1
Query: 50 SPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVD 109
+PPGYVGY EGGQLTE VRRRPY+VVLFDEIEKAH DVFN++LQ+L+DGR+TDS+GRTV
Sbjct: 1 APPGYVGYEEGGQLTEVVRRRPYSVVLFDEIEKAHHDVFNILLQLLDDGRITDSQGRTVS 180
Query: 110 FKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRL 169
F N ++IMTSN+GS I R D+K + Y+++K V E +Q F PEF+NR+
Sbjct: 181 FTNCVVIMTSNIGSHYILDTLRS-----TQDDKTAVYDQMKRQVVELARQTFHPEFMNRI 345
Query: 170 DEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPL 229
DE IVF+ L ++ +I ++ ++ V +RLK K+IDL TE+ + G+DP++GARP+
Sbjct: 346 DEYIVFQPLDSEQISKIVELQMERVKNRLKQKKIDLHYTEKAVKLLGVLGFDPNFGARPV 525
Query: 230 RRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLNGSSGAP 276
+R I +L+E+ +A +L G+ KE DS+I+DAD + L+G +P
Sbjct: 526 KRVIQQLVENEIAMGVLRGDFKEEDSIIVDAD-----VTLSGKERSP 651
>TC228467 weakly similar to UP|ERD1_ARATH (P42762) ERD1 protein, chloroplast
precursor, partial (17%)
Length = 799
Score = 162 bits (411), Expect = 1e-40
Identities = 82/164 (50%), Positives = 121/164 (73%), Gaps = 1/164 (0%)
Frame = +1
Query: 108 VDFKNTLLIMTSNVGSSVIEKGGRK-IGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFL 166
V FKN L++MTSNVGSS I KG IGF L D+K +SYN +KS+V EEL+ YFRPE L
Sbjct: 1 VSFKNALVVMTSNVGSSAITKGRHNSIGF-LIPDDKKTSYNGLKSMVIEELRTYFRPELL 177
Query: 167 NRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGA 226
NR+DE++VF+ L K ++ +I D++L+++ R+ + I + V+E ++ V ++GY+P+YGA
Sbjct: 178 NRIDEVVVFQPLEKSQLLQILDVLLQDMKKRVLSLGIHVKVSEAVKNLVCQQGYNPTYGA 357
Query: 227 RPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLN 270
RPLRRAI L+ED ++E +L GE K+GD+V++D D++G V N
Sbjct: 358 RPLRRAITSLIEDPLSEALLYGECKQGDTVLVDLDANGNPFVTN 489
>AW200876 weakly similar to GP|4105131|gb| ClpC protease {Spinacia oleracea},
partial (12%)
Length = 487
Score = 146 bits (368), Expect = 1e-35
Identities = 85/147 (57%), Positives = 99/147 (66%)
Frame = +2
Query: 28 EAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDV 87
E +I LDM M+ H+ +LI +P GY GG L EAVRR YTV +I+ AHP V
Sbjct: 47 EGIILLDMCGDMDGHSGLQLIDTPTGYSASYVGG*LLEAVRRHTYTV*RICDIDNAHPAV 226
Query: 88 FNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYN 147
FNM LQILE+GRL+DS + DF NTLLIMTSN G V EK GR I FDLDYDEKDS
Sbjct: 227 FNMKLQILEEGRLSDSLR*SEDFTNTLLIMTSNAGGRVHEKRGRII*FDLDYDEKDSMLY 406
Query: 148 RIKSLVTEELKQYFRPEFLNRLDEMIV 174
RI S VT+ELK + PE LNR ++MIV
Sbjct: 407 RIMS*VTDELKPHCMPESLNRENDMIV 487
>AW568757 similar to SP|P35100|CLPA_ ATP-dependent clp protease ATP-binding
subunit clpA homolog chloroplast precursor. [Garden
pea], partial (9%)
Length = 271
Score = 140 bits (352), Expect = 9e-34
Identities = 71/90 (78%), Positives = 80/90 (88%)
Frame = +2
Query: 194 VFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEG 253
VF RLK KEIDLSVTERFR+RVV+EGY+PSYGARPL RAIM+LLEDSMAEKMLA EI EG
Sbjct: 2 VFQRLKAKEIDLSVTERFRERVVDEGYNPSYGARPLTRAIMQLLEDSMAEKMLAREI*EG 181
Query: 254 DSVIIDADSDGKVIVLNGSSGAPESLPEAL 283
DSVI+D+DS+G VIVLNG GAP+SL + L
Sbjct: 182 DSVIVDSDSEGNVIVLNGMFGAPDSLEDVL 271
>TC225571 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (4%)
Length = 654
Score = 60.5 bits (145), Expect = 9e-10
Identities = 29/38 (76%), Positives = 34/38 (89%)
Frame = +3
Query: 246 LAGEIKEGDSVIIDADSDGKVIVLNGSSGAPESLPEAL 283
LA EIKEGDSVI+D+DS+G VIVLNGSSGAP+SL + L
Sbjct: 3 LAREIKEGDSVIVDSDSEGNVIVLNGSSGAPDSLEDVL 116
>TC218089
Length = 1393
Score = 52.4 bits (124), Expect = 2e-07
Identities = 34/127 (26%), Positives = 65/127 (50%), Gaps = 1/127 (0%)
Frame = +2
Query: 4 FSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQL 63
F G +GK ++A +LA +GS E+ I +D+S ++ G + G T +
Sbjct: 77 FVGHDRLGKKKIAVSLAELLYGSRESFIFVDLSS-------EEMKGCDVKFRGKTALDFI 235
Query: 64 TEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMT-SNVG 122
++P +VV + +EKA N + ++ G+++DS GR V NT+ + + S+
Sbjct: 236 VGECCKKPLSVVFLENVEKADILAQNSLSLAIKTGKISDSHGREVSVNNTMFVFSFSDYQ 415
Query: 123 SSVIEKG 129
+S++ +G
Sbjct: 416 NSLMPRG 436
>CD391742 homologue to GP|15912291|gb| AT4g33680/T16L1_170 {Arabidopsis
thaliana}, partial (14%)
Length = 640
Score = 50.8 bits (120), Expect = 7e-07
Identities = 23/27 (85%), Positives = 24/27 (88%)
Frame = -1
Query: 258 IDADSDGKVIVLNGSSGAPESLPEALP 284
+D DSDG VIVLNGSSGAPESLPE LP
Sbjct: 640 VDVDSDGNVIVLNGSSGAPESLPETLP 560
>TC227091 similar to UP|Q7Q6L1 (Q7Q6L1) AgCP5440 (Fragment), partial (58%)
Length = 1610
Score = 33.1 bits (74), Expect = 0.15
Identities = 37/190 (19%), Positives = 76/190 (39%), Gaps = 14/190 (7%)
Frame = +2
Query: 45 SKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSK 104
S+L G+ PG ++ + V+ Y+V++FD H +LE G
Sbjct: 596 SELAGAIPGIDEAMSFAEMLKLVQTMDYSVIVFDTAPTGHTLRLLQFPSVLEKGL----- 760
Query: 105 GRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSL--VTEELKQYFR 162
+ + KN + + R G D+ + D R++ + V E++ + F+
Sbjct: 761 AKVMSLKNKF--------GGLFNQMTRMFGMGDDFGD-DQILGRLEGMKDVIEQVNKQFK 913
Query: 163 ------------PEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTER 210
PEFL+ + + ++LTK E+ I+ + +FD + L +
Sbjct: 914 DPDMTTFVCVCIPEFLSLYETERLVQELTKFEIDTHNIIINQVIFDDEDVESKLLKARMK 1093
Query: 211 FRDRVVEEGY 220
+ + +++ Y
Sbjct: 1094MQQKYLDQFY 1123
>TC209114 similar to GB|AAP21288.1|30102740|BT006480 At1g52360 {Arabidopsis
thaliana;} , partial (22%)
Length = 849
Score = 32.7 bits (73), Expect = 0.19
Identities = 19/73 (26%), Positives = 36/73 (49%)
Frame = +3
Query: 205 LSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDG 264
+++ E FR+ +EEG P + E+ E+ GE + ++V++DADS
Sbjct: 171 ITLVEAFRNMQIEEGDQPLENGDSNHELTEQNGEEHYTEEQ-NGEGSQEEAVVVDADSTD 347
Query: 265 KVIVLNGSSGAPE 277
+++NG+ E
Sbjct: 348 GAVLVNGNEADEE 386
>TC205800 similar to GB|AAP21288.1|30102740|BT006480 At1g52360 {Arabidopsis
thaliana;} , partial (52%)
Length = 1234
Score = 30.8 bits (68), Expect = 0.73
Identities = 19/76 (25%), Positives = 36/76 (47%), Gaps = 3/76 (3%)
Frame = +2
Query: 205 LSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEG---DSVIIDAD 261
+++ E FR+ +EEG + R E+ E+ +EG ++V++DAD
Sbjct: 626 ITLVEAFRNMQIEEGEEHLENGDSTHELTERNGEEHYTEEQEEQNGEEGSQEEAVVVDAD 805
Query: 262 SDGKVIVLNGSSGAPE 277
S +++NG+ E
Sbjct: 806 STDGAVLVNGNEADEE 853
>TC204689 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATPase,
partial (98%)
Length = 1726
Score = 30.4 bits (67), Expect = 0.95
Identities = 24/81 (29%), Positives = 36/81 (43%)
Frame = +3
Query: 1 SFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEG 60
+F+ GP G GKS LAK +A+ +E + S+ VSK +G V
Sbjct: 678 AFLLYGPPGTGKSYLAKAVAT---EAESTFFSVSSSDL-----VSKWMGESEKLV----- 818
Query: 61 GQLTEAVRRRPYTVVLFDEIE 81
L E R +++ DEI+
Sbjct: 819 SNLFEMARESAPSIIFIDEID 881
>TC217900 UP|O48924 (O48924) CYP83D1p (Fragment), complete
Length = 1672
Score = 29.6 bits (65), Expect = 1.6
Identities = 24/82 (29%), Positives = 41/82 (49%), Gaps = 2/82 (2%)
Frame = +2
Query: 182 EVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSM 241
EVK+I DI+L+ + DR T ++ L + + G DPS + + A+ LL++
Sbjct: 839 EVKDIIDILLQLLDDRSFTFDLTLDHIKAVLMNIFIAGTDPS--SATIVWAMNALLKNPN 1012
Query: 242 AEKMLAGEIKE--GDSVIIDAD 261
+ GE++ GD I+ D
Sbjct: 1013VMSKVQGEVRNLFGDKDFINED 1078
>TC209041 weakly similar to UP|Q9V780 (Q9V780) Lap1 protein, partial (4%)
Length = 1041
Score = 28.9 bits (63), Expect = 2.8
Identities = 21/63 (33%), Positives = 33/63 (52%), Gaps = 2/63 (3%)
Frame = +1
Query: 92 LQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSV--IEKGGRKIGFDLDYDEKDSSYNRI 149
L+IL D RL + D +N + + T NV + ++ +GF L E D SYN+I
Sbjct: 358 LRIL-DARLNCLRSLPEDLENLINLETLNVSQNFQYLDSLPYSVGFLLSLVELDVSYNKI 534
Query: 150 KSL 152
++L
Sbjct: 535 RAL 543
>TC216063 UP|C822_SOYBN (O81972) Cytochrome P450 82A2 (P450 CP4) , complete
Length = 1766
Score = 27.7 bits (60), Expect = 6.2
Identities = 28/118 (23%), Positives = 48/118 (39%), Gaps = 7/118 (5%)
Frame = +2
Query: 139 YDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIV-------FRQLTKLEVKEIADIML 191
Y D + + + L++ L Y R MIV +RQL K+ + E
Sbjct: 305 YTTNDIAVSSLPDLISANLLCYNR--------SMIVVAPYGPYWRQLRKILMSEFLSPSR 460
Query: 192 KEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGE 249
E ++ E+ S+TE FRD + + L++ LL +M +M+ G+
Sbjct: 461 VEQLHHVRVSEVQSSITELFRDWRSNKNVQSGFATVELKQ-WFSLLVFNMILRMVCGK 631
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.137 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,964,530
Number of Sequences: 63676
Number of extensions: 77558
Number of successful extensions: 407
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 401
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 401
length of query: 284
length of database: 12,639,632
effective HSP length: 96
effective length of query: 188
effective length of database: 6,526,736
effective search space: 1227026368
effective search space used: 1227026368
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0266.20


