

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
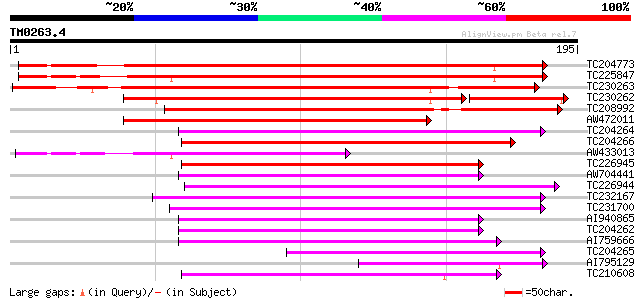
Query= TM0263.4
(195 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204773 UP|OLE1_SOYBN (P29530) P24 oleosin isoform A (P89), com... 183 4e-47
TC225847 UP|OLE2_SOYBN (P29531) P24 oleosin isoform B (P91), com... 178 1e-45
TC230263 weakly similar to UP|OLE1_SOYBN (P29530) P24 oleosin is... 176 8e-45
TC230262 weakly similar to UP|Q6J1J8 (Q6J1J8) Oleosin isoform, p... 112 8e-34
TC208992 weakly similar to UP|O04925 (O04925) 15.5 kDa oleosin, ... 110 5e-25
AW472011 similar to SP|P29530|OLE1 P24 oleosin isoform A (P89). ... 102 1e-22
TC204264 similar to UP|Q8LL70 (Q8LL70) 15.8 kDa oleosin, partial... 96 1e-20
TC204266 similar to UP|Q8LL70 (Q8LL70) 15.8 kDa oleosin, partial... 91 3e-19
AW433013 similar to SP|P29531|OLE2 P24 oleosin isoform B (P91). ... 89 1e-18
TC226945 similar to UP|OLE1_PRUDU (Q43804) Oleosin 1, partial (72%) 86 1e-17
AW704441 similar to GP|21311555|gb| 15.8 kDa oleosin {Theobroma ... 84 4e-17
TC226944 similar to UP|OLE1_PRUDU (Q43804) Oleosin 1, partial (78%) 84 5e-17
TC232167 similar to PIR|E96527|E96527 protein F27J15.22 [importe... 80 6e-16
TC231700 weakly similar to PIR|E96527|E96527 protein F27J15.22 [... 80 6e-16
AI940865 weakly similar to GP|21311555|gb| 15.8 kDa oleosin {The... 77 5e-15
TC204262 weakly similar to UP|Q8LL70 (Q8LL70) 15.8 kDa oleosin, ... 76 9e-15
AI759666 weakly similar to GP|21311555|gb 15.8 kDa oleosin {Theo... 75 1e-14
TC204265 weakly similar to UP|Q8LL70 (Q8LL70) 15.8 kDa oleosin, ... 68 3e-12
AI795129 similar to SP|P29531|OLE2_ P24 oleosin isoform B (P91).... 61 4e-10
TC210608 weakly similar to PIR|E96527|E96527 protein F27J15.22 [... 58 3e-09
>TC204773 UP|OLE1_SOYBN (P29530) P24 oleosin isoform A (P89), complete
Length = 1048
Score = 183 bits (465), Expect = 4e-47
Identities = 102/189 (53%), Positives = 124/189 (64%), Gaps = 7/189 (3%)
Frame = +3
Query: 4 PQQQVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLFPERGPSASQ 63
P V VHTT THRYE G P R+E P RY G S ERGP+ SQ
Sbjct: 102 PPHSVQVHTT-THRYEAGVVPPGARFE---------PPRYEAGVKAPSIYHSERGPTTSQ 251
Query: 64 IIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILTS 123
++AV AG+P+GG LLLLAG++L +L GLAV TPL +LFSPV+VPA + IGLAVAG LTS
Sbjct: 252 VLAVLAGLPVGGILLLLAGLTLAGTLTGLAVATPLFVLFSPVLVPATVAIGLAVAGFLTS 431
Query: 124 GAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADY-------VGQKTKEVGHD 176
GAFGLT L SFSW++NYIRET+ AK+ +A+ A+Y VGQKTKEVG D
Sbjct: 432 GAFGLTALSSFSWILNYIRETQPASENLAAAAKHHLAEAAEYVGQKTKEVGQKTKEVGQD 611
Query: 177 IQTKAHEAK 185
IQ+KA + +
Sbjct: 612 IQSKAQDTR 638
>TC225847 UP|OLE2_SOYBN (P29531) P24 oleosin isoform B (P91), complete
Length = 1027
Score = 178 bits (452), Expect = 1e-45
Identities = 101/190 (53%), Positives = 126/190 (66%), Gaps = 8/190 (4%)
Frame = +2
Query: 4 PQQQVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLF-PERGPSAS 62
P V VHTT THRYE G PP R+EA RY G SS++ ERGP+ S
Sbjct: 119 PPHSVQVHTT-THRYEAGVV-PPARFEA---------PRYEAGIKAPSSIYHSERGPTTS 265
Query: 63 QIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGILT 122
Q++AV AG+P+GG LLLLAG++L +L GL V TPL I+FSPV++PA + IGLAVAG LT
Sbjct: 266 QVLAVVAGLPVGGILLLLAGLTLAGTLTGLVVATPLFIIFSPVLIPATVAIGLAVAGFLT 445
Query: 123 SGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADY-------VGQKTKEVGH 175
SG FGLT L SFSW++NYIRET+ AK+ +A+ A+Y VGQKTKEVG
Sbjct: 446 SGVFGLTALSSFSWILNYIRETQPASENLAAAAKHHLAEAAEYVGQKTKEVGQKTKEVGQ 625
Query: 176 DIQTKAHEAK 185
DIQ+KA + +
Sbjct: 626 DIQSKAQDTR 655
>TC230263 weakly similar to UP|OLE1_SOYBN (P29530) P24 oleosin isoform A
(P89), partial (40%)
Length = 1004
Score = 176 bits (445), Expect = 8e-45
Identities = 100/190 (52%), Positives = 126/190 (65%), Gaps = 9/190 (4%)
Frame = +2
Query: 2 AQPQQQVHVHTTATHRYETGGANPPQ-RYEAGGINFNNQPQRYGGGGGGFSSLFPERGPS 60
+Q Q V VH + TH P Q RY GG + Q +GG GGG SLFPE +
Sbjct: 116 SQQPQHVQVHASTTH-------TPQQYRYYQGGA----KTQHHGGEGGGVMSLFPEISLT 262
Query: 61 ASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGI 120
SQ++A+ AGVP+GG LLLL+G+SLIASL+GLAV TPL I FSPV+VPA IG+AV +
Sbjct: 263 GSQLLALLAGVPLGGMLLLLSGVSLIASLVGLAVATPLFIFFSPVLVPAAFVIGMAVTAV 442
Query: 121 LTSGAFGLTGLMSFSWLMNYIRE--------TRGIVPEQLDYAKNRMADMADYVGQKTKE 172
L +GA GL GL+SFSWL+N +R+ T + PEQ AK +ADM +YVG+KTK+
Sbjct: 443 LAAGACGLVGLVSFSWLVNCLRQMPRGTTTKTTMMRPEQ---AKRHVADMEEYVGKKTKD 613
Query: 173 VGHDIQTKAH 182
VG DIQT+AH
Sbjct: 614 VGQDIQTRAH 643
>TC230262 weakly similar to UP|Q6J1J8 (Q6J1J8) Oleosin isoform, partial (47%)
Length = 716
Score = 112 bits (280), Expect(2) = 8e-34
Identities = 61/125 (48%), Positives = 84/125 (66%), Gaps = 7/125 (5%)
Frame = +2
Query: 40 PQRYGGGGGG-FSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPL 98
PQ +GG GG +L PE + SQ++A+ AGVP+ LLLL+GISLIASL+GLAV TPL
Sbjct: 38 PQHHGGEGGSRILNLIPEMSLTGSQLLALLAGVPLVAMLLLLSGISLIASLLGLAVATPL 217
Query: 99 LILFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRE------TRGIVPEQL 152
I FSP++VPA IG+AV +L + A L GL+ FSW++NY+R+ ++PEQ
Sbjct: 218 YIFFSPLLVPAAFAIGMAVTAVLAAEACRLAGLVLFSWVVNYLRQMPRRTTMMTVLPEQA 397
Query: 153 DYAKN 157
A++
Sbjct: 398 IAARS 412
Score = 48.1 bits (113), Expect(2) = 8e-34
Identities = 23/35 (65%), Positives = 29/35 (82%), Gaps = 1/35 (2%)
Frame = +3
Query: 159 MADMADYVGQKTKEVGHDIQTKAHEAKRTP-ITTH 192
+ADMA+YVGQKTK+V DIQT+AH A+RT +T H
Sbjct: 408 VADMAEYVGQKTKDVRQDIQTRAHHAQRTTRMTVH 512
>TC208992 weakly similar to UP|O04925 (O04925) 15.5 kDa oleosin, partial
(55%)
Length = 937
Score = 110 bits (274), Expect = 5e-25
Identities = 60/137 (43%), Positives = 84/137 (60%)
Frame = +1
Query: 54 FPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTI 113
+ +R PS S ++ +AA VP G +LL+L G++L A++I L V TPL ++FSPV++PA + I
Sbjct: 55 YMKRKPSNSNVLVLAALVPFGVSLLILVGLTLSATVISLTVVTPLFVIFSPVLLPAALFI 234
Query: 114 GLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEV 173
LAVAG LTSG FG+T + SF+WL +R + PE R D AD V QKT E
Sbjct: 235 ALAVAGFLTSGVFGITSISSFAWLATNLRRSHS--PEH----PQRPRDDADRVAQKTDEA 396
Query: 174 GHDIQTKAHEAKRTPIT 190
Q H+A++ T
Sbjct: 397 VRQAQETVHQAQKETAT 447
>AW472011 similar to SP|P29530|OLE1 P24 oleosin isoform A (P89). [Soybean]
{Glycine max}, partial (50%)
Length = 418
Score = 102 bits (254), Expect = 1e-22
Identities = 52/106 (49%), Positives = 70/106 (65%)
Frame = +2
Query: 40 PQRYGGGGGGFSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLL 99
P RY G G S E+GPSA ++ G+P+GG LLLLAG++L +L GLA+ P+
Sbjct: 92 PPRYEAGVGAPSIYSFEKGPSAFHVLGRLTGLPVGGILLLLAGLTLAGTLTGLAIARPIC 271
Query: 100 ILFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETR 145
LFSP++VPA++ I L+ T GAF LT L FSW++NYIRET+
Sbjct: 272 RLFSPMLVPAMVAISLSEIRFQTYGAFRLTALAFFSWILNYIRETQ 409
>TC204264 similar to UP|Q8LL70 (Q8LL70) 15.8 kDa oleosin, partial (82%)
Length = 853
Score = 95.9 bits (237), Expect = 1e-20
Identities = 47/126 (37%), Positives = 74/126 (58%)
Frame = +3
Query: 59 PSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVA 118
P + Q + IG TLLLL+G++L ++IGL + TPLL++FSP++VPA + L +
Sbjct: 180 PPSRQTVKFITAATIGITLLLLSGLTLTGTVIGLIIATPLLVIFSPILVPAAFVLFLVAS 359
Query: 119 GILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGHDIQ 178
G L SG G+ + + SW+ NY+ + + LDYAK + D A V ++ K+ G Q
Sbjct: 360 GFLFSGGCGVAAIAALSWIYNYVSGNQPAGSDTLDYAKGYLTDKARDVKERAKDYGSYAQ 539
Query: 179 TKAHEA 184
+ +EA
Sbjct: 540 GRINEA 557
>TC204266 similar to UP|Q8LL70 (Q8LL70) 15.8 kDa oleosin, partial (80%)
Length = 754
Score = 91.3 bits (225), Expect = 3e-19
Identities = 44/115 (38%), Positives = 70/115 (60%)
Frame = +2
Query: 60 SASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAG 119
++ Q + IG TLLLL+G++L ++IGL + TPLL++FSP++VPA + L +G
Sbjct: 11 ASRQTVKFITAATIGITLLLLSGLTLTGTVIGLIIATPLLVIFSPILVPAAFVLFLVASG 190
Query: 120 ILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVG 174
L SG G+ + + SW+ NY+ + + LDYAK +AD A V ++ K+ G
Sbjct: 191 FLFSGGCGVAAIAALSWIYNYVSGNQPAGYDTLDYAKGYLADKARDVKERAKDYG 355
>AW433013 similar to SP|P29531|OLE2 P24 oleosin isoform B (P91). [Soybean]
{Glycine max}, partial (47%)
Length = 325
Score = 89.4 bits (220), Expect = 1e-18
Identities = 54/116 (46%), Positives = 70/116 (59%), Gaps = 1/116 (0%)
Frame = +2
Query: 3 QPQQQVHVHTTATHRYETGGANPPQRYEAGGINFNNQPQRYGGGGGGFSSLF-PERGPSA 61
+P V HTT THRYE G PP R EA Y G SS++ ERGP+
Sbjct: 8 EPPHSVQGHTT-THRYEAGVV-PPARLEAPS---------YEAWIGAPSSIYHAERGPTT 154
Query: 62 SQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAV 117
Q++ VAAG+P+ G LLLLAG++L +L L V TP+ I+FSPV++P + IGLAV
Sbjct: 155 CQVLTVAAGLPVCGILLLLAGLTLTGTLTRLVVVTPIFIIFSPVLIPDAVAIGLAV 322
>TC226945 similar to UP|OLE1_PRUDU (Q43804) Oleosin 1, partial (72%)
Length = 766
Score = 85.9 bits (211), Expect = 1e-17
Identities = 40/104 (38%), Positives = 64/104 (61%)
Frame = +3
Query: 60 SASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAG 119
S Q++ A V GG+LL+LA + L A++I L + TPL ++FSPV+VPAV+T+ L G
Sbjct: 132 STHQVVKAATAVTAGGSLLILASLILAATVIALTIVTPLFVIFSPVLVPAVITVALLSLG 311
Query: 120 ILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
L S FG+ + +W+ Y+ + +QLD A++++ D A
Sbjct: 312 FLASSGFGVAAITVLAWIYRYVTGKQPPGADQLDSARHKIMDKA 443
>AW704441 similar to GP|21311555|gb| 15.8 kDa oleosin {Theobroma cacao},
partial (65%)
Length = 337
Score = 84.0 bits (206), Expect = 4e-17
Identities = 42/105 (40%), Positives = 62/105 (59%)
Frame = +2
Query: 59 PSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVA 118
PS + V IG TLLLL+G++L ++IGL + TPLL +FSP++VPA + L +
Sbjct: 17 PSCTPDREVITAATIGSTLLLLSGLTLTGTVIGLIIATPLLDIFSPILVPAAFVLFLVAS 196
Query: 119 GILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
G L SG G+ + + SW+ NY+ + + LDYAK + D A
Sbjct: 197 GFLFSGG*GVAAIAALSWIYNYVSGYQPAESDTLDYAKGYLTDNA 331
>TC226944 similar to UP|OLE1_PRUDU (Q43804) Oleosin 1, partial (78%)
Length = 758
Score = 83.6 bits (205), Expect = 5e-17
Identities = 43/129 (33%), Positives = 72/129 (55%)
Frame = +3
Query: 61 ASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAGI 120
++Q++ A V GG+LL+LA + L ++I L + TP L++FSPV+VPAV+T+ L G
Sbjct: 135 STQVVKAATAVTAGGSLLILASLILAGTIIALTIVTPPLVIFSPVLVPAVITVALLSLGF 314
Query: 121 LTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGHDIQTK 180
L SG FG+ + +W+ Y+ +QLD A +++ D A + ++ +Q
Sbjct: 315 LASGGFGVAAITVLAWIYRYVTGKYPPGADQLDSAPHKIMDKAREIKDYGQQQISGVQAS 494
Query: 181 AHEAKRTPI 189
A+ PI
Sbjct: 495 *SCARTHPI 521
>TC232167 similar to PIR|E96527|E96527 protein F27J15.22 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(30%)
Length = 740
Score = 80.1 bits (196), Expect = 6e-16
Identities = 41/135 (30%), Positives = 74/135 (54%)
Frame = +3
Query: 50 FSSLFPERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPA 109
F +R P+++Q+ + + LLL G+++ +++GL F PL+I+ SPV VPA
Sbjct: 117 FLQKLQDRAPNSTQLAGILPLLVTASVFLLLTGLTVAGAVLGLIFFLPLIIVSSPVWVPA 296
Query: 110 VMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQK 169
+ + AG L+ FG+ + + SW+ Y R +++DYA++R+ D A +V
Sbjct: 297 GTLLFIVTAGFLSVCGFGVALVAALSWMYRYFRGLHPPGSDRVDYARSRIYDTASHVKDY 476
Query: 170 TKEVGHDIQTKAHEA 184
+E G +Q+K +A
Sbjct: 477 AREYGGYLQSKVKDA 521
>TC231700 weakly similar to PIR|E96527|E96527 protein F27J15.22 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(58%)
Length = 752
Score = 80.1 bits (196), Expect = 6e-16
Identities = 41/129 (31%), Positives = 74/129 (56%)
Frame = +2
Query: 56 ERGPSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGL 115
+RGP+++Q+ + + LLLL G+++ +++GL F PL+I+ SPV VPA + +
Sbjct: 134 DRGPNSTQLACILTLLLTASVLLLLTGLTVAGAVLGLIFFLPLIIVSSPVWVPAGTFLFI 313
Query: 116 AVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQKTKEVGH 175
A L+ FG+ + SW+ +Y R +++DYA++R+ D A +V +E G
Sbjct: 314 VTAAFLSVCGFGVALVAVLSWMYHYFRGLHPPGSDRVDYARSRIYDTASHVKDYAREYGG 493
Query: 176 DIQTKAHEA 184
+Q+K +A
Sbjct: 494 YLQSKVKDA 520
>AI940865 weakly similar to GP|21311555|gb| 15.8 kDa oleosin {Theobroma
cacao}, partial (55%)
Length = 319
Score = 77.0 bits (188), Expect = 5e-15
Identities = 39/105 (37%), Positives = 60/105 (57%)
Frame = +2
Query: 59 PSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVA 118
P++ + IG TLLL +G +L ++IGL + TPLL++FSP++ PA + + L +
Sbjct: 5 PTSRPTVKCITAATIGITLLLQSG*TLTGTVIGLIIATPLLVIFSPILAPAALVLLLVAS 184
Query: 119 GILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
G L SG G + + SW+ NY + + LDYAK +AD A
Sbjct: 185 GFLFSGGCGEAAIAA*SWIHNYESANQSATYDTLDYAKRYLADNA 319
>TC204262 weakly similar to UP|Q8LL70 (Q8LL70) 15.8 kDa oleosin, partial
(73%)
Length = 444
Score = 76.3 bits (186), Expect = 9e-15
Identities = 37/105 (35%), Positives = 60/105 (56%)
Frame = +1
Query: 59 PSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVA 118
P + Q + IG TLLLL+G++L + IG + T +L++F P++VPA + + L +
Sbjct: 121 PPSRQTVKFITAATIGITLLLLSGLTLTGTDIGSIIATTVLVIFRPILVPAALVLLLVAS 300
Query: 119 GILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMA 163
G L SG G+ + + SW+ NY+ + + LDYAK + D A
Sbjct: 301 GYLLSGGCGVAAIAALSWIYNYVFGNQPAGSDTLDYAKGYLTDNA 435
>AI759666 weakly similar to GP|21311555|gb 15.8 kDa oleosin {Theobroma
cacao}, partial (76%)
Length = 343
Score = 75.5 bits (184), Expect = 1e-14
Identities = 37/111 (33%), Positives = 59/111 (52%)
Frame = +2
Query: 59 PSASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVA 118
P + Q + IG TLLLL+G++L ++IGL + P+L++ SP++VPA + L
Sbjct: 5 PPSRQTVKFITDATIGITLLLLSGLTLTGTVIGLIIAAPILVILSPILVPAAFVLCLVAT 184
Query: 119 GILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVGQK 169
G L S + + SW NY+ + + LDYAK + D A V ++
Sbjct: 185 GFLFSAGCDEAAIAALSWTYNYVSANQPAGSDTLDYAKGYLTDKARDVNER 337
>TC204265 weakly similar to UP|Q8LL70 (Q8LL70) 15.8 kDa oleosin, partial
(52%)
Length = 572
Score = 67.8 bits (164), Expect = 3e-12
Identities = 32/89 (35%), Positives = 51/89 (56%)
Frame = +2
Query: 96 TPLLILFSPVIVPAVMTIGLAVAGILTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYA 155
TP+L LFSP++VPA + L +G + SG G+ + + SW+ NY+ + + LDYA
Sbjct: 5 TPVLALFSPILVPAAFVLLLTASGFVLSGGCGVAAIAALSWIYNYVSGNQPAGSDTLDYA 184
Query: 156 KNRMADMADYVGQKTKEVGHDIQTKAHEA 184
K + D A V ++ K+ G Q + +EA
Sbjct: 185 KGYLTDKARDVKERAKDYGSYAQGRINEA 271
>AI795129 similar to SP|P29531|OLE2_ P24 oleosin isoform B (P91). [Soybean]
{Glycine max}, partial (32%)
Length = 217
Score = 60.8 bits (146), Expect = 4e-10
Identities = 33/72 (45%), Positives = 43/72 (58%), Gaps = 7/72 (9%)
Frame = +2
Query: 121 LTSGAFGLTGLMSFSWLMNYIRETRGIVPEQLDYAKNRMADMADYVG-------QKTKEV 173
LTS FGLT L FSW++NYIR T+ +K+ +AD A+YVG QKT EV
Sbjct: 2 LTSEVFGLTALSFFSWILNYIR*TQPASENLAAASKHHLADAAEYVGHKTKEVWQKTNEV 181
Query: 174 GHDIQTKAHEAK 185
G DI +KA + +
Sbjct: 182 GQDIHSKAQDTR 217
>TC210608 weakly similar to PIR|E96527|E96527 protein F27J15.22 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(29%)
Length = 589
Score = 57.8 bits (138), Expect = 3e-09
Identities = 32/111 (28%), Positives = 58/111 (51%), Gaps = 1/111 (0%)
Frame = +2
Query: 60 SASQIIAVAAGVPIGGTLLLLAGISLIASLIGLAVFTPLLILFSPVIVPAVMTIGLAVAG 119
+++Q+ V + G LLLL G +++ S++GL +F PL+I+ SP+ +P + L A
Sbjct: 116 NSNQLFGVLTLLITGSILLLLTGPTIVGSVLGLILFAPLIIVTSPIWIPLCAVLFLVTAM 295
Query: 120 ILTSGAFGLTGLMSFSWLMNYIRETRGIV-PEQLDYAKNRMADMADYVGQK 169
L+ FG+ L +W+ Y + ++ +A NR+ D A + K
Sbjct: 296 FLSMCGFGIVVLAVLTWMYRYFKGLHHPPGSNRVSHALNRVKDRAGNLNSK 448
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,482,610
Number of Sequences: 63676
Number of extensions: 98546
Number of successful extensions: 911
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 838
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 879
length of query: 195
length of database: 12,639,632
effective HSP length: 92
effective length of query: 103
effective length of database: 6,781,440
effective search space: 698488320
effective search space used: 698488320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0263.4


