

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
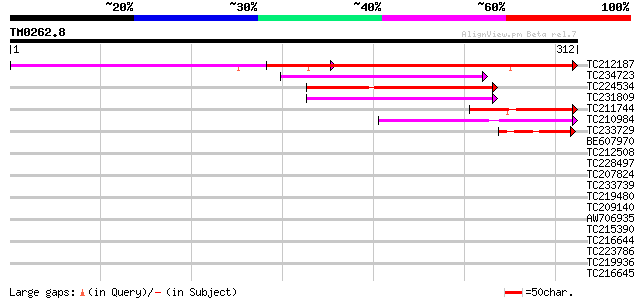
Query= TM0262.8
(312 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC212187 weakly similar to UP|Q6USA0 (Q6USA0) Transposase (Fragm... 214 4e-56
TC234723 similar to UP|Q9FHY5 (Q9FHY5) Emb|CAB72466.1 (At5g41980... 96 2e-20
TC224534 similar to UP|Q6USA0 (Q6USA0) Transposase (Fragment), p... 95 4e-20
TC231809 84 8e-17
TC211744 weakly similar to UP|O46921 (O46921) Thylakoid-bound as... 75 5e-14
TC210984 71 7e-13
TC233729 47 1e-05
BE607970 37 0.009
TC212508 35 0.044
TC228497 33 0.13
TC207824 similar to GB|AAN73294.1|25141199|BT002297 At5g12010/F1... 33 0.17
TC233739 32 0.28
TC219480 similar to UP|Q6UEI6 (Q6UEI6) Early flowering 4, partia... 29 3.1
TC209140 weakly similar to UP|Q948R9 (Q948R9) RPS6-like protein ... 28 4.1
AW706935 homologue to GP|15809840|gb| AT4g30400/F17I23_260 {Arab... 28 4.1
TC215390 similar to UP|Q9ZR83 (Q9ZR83) EREBP-3 homolog, partial ... 28 5.3
TC216644 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [... 28 7.0
TC223786 similar to UP|Q8I738 (Q8I738) TcC31.4, partial (11%) 28 7.0
TC219936 similar to UP|Q9LJL8 (Q9LJL8) Emb|CAB43653.1 (AT3g19120... 27 9.1
TC216645 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [... 27 9.1
>TC212187 weakly similar to UP|Q6USA0 (Q6USA0) Transposase (Fragment),
partial (45%)
Length = 973
Score = 214 bits (545), Expect = 4e-56
Identities = 107/178 (60%), Positives = 132/178 (74%), Gaps = 7/178 (3%)
Frame = +1
Query: 142 *NL*LYNIVTLMY*HAIIFYCL---GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPR 198
*NL + NI ++ + + YC +YLVD+ YPTFMGFLG Y K RYHLPQFR GPR
Sbjct: 433 *NLYIINICIIIILCSNMLYCFIL*VNFYLVDSCYPTFMGFLGSYNKTRYHLPQFRIGPR 612
Query: 199 ITGRVEVFNYYHSSLRNVIERSFGCCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRR 258
I GRVEVFNYYHSSL++ IER+FG CKA+WKILG+M + LKTQN+II ACM +HNFI+R
Sbjct: 613 IRGRVEVFNYYHSSLQSTIERAFGLCKARWKILGNMSPFALKTQNQIIVACMTIHNFIQR 792
Query: 259 NDRSDEEFDANYENED----GGGENEAGPSTVTWEEPDFQSSLQMEQIREHIKNMFPT 312
ND+SD EFD+ E+ + E+E GPST TWEE D QS+L ME+ RE +KN+FPT
Sbjct: 793 NDKSDGEFDSLDEDNEYIDTDKDESEVGPSTTTWEESDAQSTLLMERFRESLKNIFPT 966
Score = 121 bits (303), Expect = 5e-28
Identities = 87/182 (47%), Positives = 103/182 (55%), Gaps = 3/182 (1%)
Frame = +3
Query: 1 LFAI*RRDSNILEKQYLDSFIAS*MLFVDLQET*LNLLTHHLVMFLMRLRMMSDITLILK 60
LF I R+D NI KQY+ +I S* L V LQ *L+LL HHL + LMR M DI LIL
Sbjct: 18 LFVIVRKDFNIQVKQYIGIYIVS*KLCVCLQRI*LSLLIHHLEILLMRF*KMLDIALILG 197
Query: 61 IALVQ*MVLT*EFVFLLNSRQLILVGKVTLPLMSWLLVISTCVSPLSGLGGRVLHMMLRF 120
I LVQ*MVL EFVF L ++ I+V K TLPLMS L V CVS L G GRVLHM+LR+
Sbjct: 198 IVLVQ*MVLIYEFVFPLIYKESIVVEKATLPLMS*LFVTLACVSLLFGQVGRVLHMILRY 377
Query: 121 LWRL---CVDHHCIFPILLMVRNI*NL*LYNIVTLMY*HAIIFYCLGKYYLVDAGYPTFM 177
C+ L + YN V +*HA++FY LGK+ P F
Sbjct: 378 FGGFT*ACIAFSTSSSRYLKFIYYKYMYHYNFV---F*HALLFYSLGKFLSC*FLLPYFY 548
Query: 178 GF 179
GF
Sbjct: 549 GF 554
>TC234723 similar to UP|Q9FHY5 (Q9FHY5) Emb|CAB72466.1 (At5g41980), partial
(24%)
Length = 837
Score = 96.3 bits (238), Expect = 2e-20
Identities = 50/114 (43%), Positives = 67/114 (57%)
Frame = +2
Query: 150 VTLMY*HAIIFYCLGKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYY 209
V Y* + Y GKYY+VD+ Y GF+ PY Y+ +F + E+FN
Sbjct: 95 VCFKY*IKTLVYDTGKYYIVDSKYQNVPGFVAPYSSTPYYSKEFLSDYHPQDASELFNQR 274
Query: 210 HSSLRNVIERSFGCCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSD 263
HS LR+VI+R+FG KA++ IL S PSY L+TQ K++ A ALHN+IRR D
Sbjct: 275 HSLLRHVIDRTFGILKARFPILMSAPSYPLQTQVKLVVAACALHNYIRREKPDD 436
>TC224534 similar to UP|Q6USA0 (Q6USA0) Transposase (Fragment), partial (50%)
Length = 968
Score = 95.1 bits (235), Expect = 4e-20
Identities = 51/106 (48%), Positives = 66/106 (62%), Gaps = 1/106 (0%)
Frame = +1
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQF-RNGPRITGRVEVFNYYHSSLRNVIERSFG 222
GKY+LVDAGY GFL PY + RYH ++ N P+ E+FN H+S +NVIERSFG
Sbjct: 475 GKYFLVDAGYTNGPGFLTPY*RTRYHRNEWIGNTPQ--NYKELFNLRHASAKNVIERSFG 648
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFDA 268
K +W IL + +D+KTQ +II C L NFIR +SD +A
Sbjct: 649 VLKKRWSILRTPSFFDIKTQIRIINTCFMLRNFIRDEQQSDPILEA 786
>TC231809
Length = 760
Score = 84.0 bits (206), Expect = 8e-17
Identities = 45/106 (42%), Positives = 60/106 (56%), Gaps = 1/106 (0%)
Frame = +3
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRV-EVFNYYHSSLRNVIERSFG 222
GKYYLV GY GF+ PY+ RY +FR ++ E+FN+ H LRN I RSF
Sbjct: 87 GKYYLVXKGYLNTEGFIAPYQGVRYQHYEFRGANQLPRNAKELFNHRHCFLRNAILRSFN 266
Query: 223 CCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSDEEFDA 268
K ++ IL P Y + Q I+ A LHNFIRR +R+D F++
Sbjct: 267 VLKTRFPILKLAPQYSFQIQRDIVIAGCVLHNFIRREERNDWIFNS 404
>TC211744 weakly similar to UP|O46921 (O46921) Thylakoid-bound ascorbate
peroxidase, partial (6%)
Length = 305
Score = 74.7 bits (182), Expect = 5e-14
Identities = 39/66 (59%), Positives = 48/66 (72%), Gaps = 7/66 (10%)
Frame = +3
Query: 254 NFIRRNDRSDEEFDANYEN-------EDGGGENEAGPSTVTWEEPDFQSSLQMEQIREHI 306
NFI+RND+SD EF++ E+ EDG +E GPST TWEEPD QS+LQME+ RE +
Sbjct: 21 NFIQRNDKSDGEFNSLDEDNADIDSDEDG---SEVGPSTTTWEEPDAQSTLQMERFRESL 191
Query: 307 KNMFPT 312
KNMFPT
Sbjct: 192 KNMFPT 209
>TC210984
Length = 1085
Score = 70.9 bits (172), Expect = 7e-13
Identities = 39/109 (35%), Positives = 61/109 (55%)
Frame = +3
Query: 204 EVFNYYHSSLRNVIERSFGCCKAKWKILGSMPSYDLKTQNKIITACMALHNFIRRNDRSD 263
E+FN H+SLRNVIER FG K+++ I S P + KTQ +++ AC ALHNF+R+ RSD
Sbjct: 426 ELFNLRHASLRNVIERIFGIFKSRFTIFKSAPPFLFKTQAELVLACAALHNFLRKECRSD 605
Query: 264 EEFDANYENEDGGGENEAGPSTVTWEEPDFQSSLQMEQIREHIKNMFPT 312
E + E + + +E+ D + +Q ++ N++ T
Sbjct: 606 E-----FPVEPTDESSSSSSVLPNYEDNDHEPIVQTQEQEREDANIWRT 737
>TC233729
Length = 580
Score = 47.0 bits (110), Expect = 1e-05
Identities = 24/42 (57%), Positives = 30/42 (71%)
Frame = +1
Query: 270 YENEDGGGENEAGPSTVTWEEPDFQSSLQMEQIREHIKNMFP 311
Y +E+ ENE GPST TW D QS+L MEQ++E IKN+FP
Sbjct: 385 YSDEE---ENEVGPSTTTW---DAQSALLMEQVKESIKNIFP 492
>BE607970
Length = 485
Score = 37.4 bits (85), Expect = 0.009
Identities = 23/95 (24%), Positives = 42/95 (44%), Gaps = 3/95 (3%)
Frame = +1
Query: 165 KYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGCC 224
+Y L D+ +P L PY + F + R FN H + ++ +FG
Sbjct: 208 QYILGDSCFPLLPWLLTPYNRVNEE-DSFGSAER------AFNCVHGNAMELVGDAFGRL 366
Query: 225 KAKWKILGSMPSYDLKTQNK---IITACMALHNFI 256
+A+W++L + + + ++ AC LHNF+
Sbjct: 367 RARWRLLAASRKWKQECVEHLPFVVVACCLLHNFV 471
>TC212508
Length = 752
Score = 35.0 bits (79), Expect = 0.044
Identities = 24/104 (23%), Positives = 41/104 (39%), Gaps = 1/104 (0%)
Frame = +2
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGC 223
G + + ++G+P L PY +T FN ++ V + +F
Sbjct: 152 GVWIVXNSGHPLMDWVLVPYTH-----------ANLTWTQHAFNEKIEEIQGVAKEAFAR 298
Query: 224 CKAKWKILGSMPSYDLKTQNKIITACMALHNFIR-RNDRSDEEF 266
K +W L L+ ++ AC LHN RN+ D+E+
Sbjct: 299 LKGRWGCLQKRTEVKLQDLPVVLGACCVLHNICEMRNEEMDDEW 430
>TC228497
Length = 779
Score = 33.5 bits (75), Expect = 0.13
Identities = 20/91 (21%), Positives = 34/91 (36%)
Frame = +2
Query: 170 DAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGCCKAKWK 229
++G+P G L PY +T FN ++++ + +F K +W
Sbjct: 221 NSGHPLMEGVLVPYTHQN-----------LTWTQHAFNQKVGEIQSIAKDAFARLKGRWS 367
Query: 230 ILGSMPSYDLKTQNKIITACMALHNFIRRND 260
L L+ ++ AC LHN D
Sbjct: 368 CLQKRTEVKLEDLPVLLGACCVLHNICEMRD 460
>TC207824 similar to GB|AAN73294.1|25141199|BT002297 At5g12010/F14F18_180
{Arabidopsis thaliana;} , partial (38%)
Length = 847
Score = 33.1 bits (74), Expect = 0.17
Identities = 23/112 (20%), Positives = 42/112 (36%), Gaps = 1/112 (0%)
Frame = +3
Query: 164 GKYYLVDAGYPTFMGFLGPYKKNRYHLPQFRNGPRITGRVEVFNYYHSSLRNVIERSFGC 223
G + + +GYP L PY + +T FN ++ V +F
Sbjct: 165 GDWIVGSSGYPLMDWVLVPYSQQN-----------LTWTQHAFNEKIGEVQKVARDAFAR 311
Query: 224 CKAKWKILGSMPSYDLKTQNKIITACMALHNFIR-RNDRSDEEFDANYENED 274
K +W L L+ ++ AC LHN + ++ D E + +++
Sbjct: 312 LKGRWSCLQKRTEVKLQDLPVVLGACCVLHNICELKGEKIDPELKVDLMDDE 467
>TC233739
Length = 562
Score = 32.3 bits (72), Expect = 0.28
Identities = 20/49 (40%), Positives = 29/49 (58%), Gaps = 3/49 (6%)
Frame = +3
Query: 259 NDRSDEEFDANYENEDGGGENEAGPST---VTWEEPDFQSSLQMEQIRE 304
N+ +EE D N E +DG E + GPST +T EE + Q +LQ ++E
Sbjct: 51 NEDEEEEDDKNKEKKDGEDEKKKGPSTLSKLTAEEAEHQ-ALQAAVLQE 194
>TC219480 similar to UP|Q6UEI6 (Q6UEI6) Early flowering 4, partial (42%)
Length = 610
Score = 28.9 bits (63), Expect = 3.1
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 2/43 (4%)
Frame = +3
Query: 185 KNRYHLPQ--FRNGPRITGRVEVFNYYHSSLRNVIERSFGCCK 225
++R HLP+ + GPR+ RV V H +R R+ GCC+
Sbjct: 333 QHRLHLPEPLVKGGPRL--RVAVLRLLHLPVRLPGRRAQGCCR 455
>TC209140 weakly similar to UP|Q948R9 (Q948R9) RPS6-like protein
(AT3g17170/K14A17_29), partial (59%)
Length = 1111
Score = 28.5 bits (62), Expect = 4.1
Identities = 11/24 (45%), Positives = 16/24 (65%)
Frame = +3
Query: 259 NDRSDEEFDANYENEDGGGENEAG 282
+D DEEFD NY+++ G + E G
Sbjct: 762 DDNDDEEFDENYDDDWDGEDEEDG 833
>AW706935 homologue to GP|15809840|gb| AT4g30400/F17I23_260 {Arabidopsis
thaliana}, partial (10%)
Length = 426
Score = 28.5 bits (62), Expect = 4.1
Identities = 20/49 (40%), Positives = 29/49 (58%)
Frame = +3
Query: 75 FLLNSRQLILVGKVTLPLMSWLLVISTCVSPLSGLGGRVLHMMLRFLWR 123
F LN R L KV+ P + +++I V +SGL LH+++RFLWR
Sbjct: 222 FSLNDRAL----KVS-PSILLIIIILAIVFFISGL----LHLLVRFLWR 341
>TC215390 similar to UP|Q9ZR83 (Q9ZR83) EREBP-3 homolog, partial (76%)
Length = 975
Score = 28.1 bits (61), Expect = 5.3
Identities = 11/31 (35%), Positives = 17/31 (54%)
Frame = -2
Query: 252 LHNFIRRNDRSDEEFDANYENEDGGGENEAG 282
+HN +RR R+ E + E E GG+ + G
Sbjct: 461 VHNLVRRRHRAGENRELVPEGESSGGDGDGG 369
>TC216644 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [Contains:
Apg8FG], partial (76%)
Length = 740
Score = 27.7 bits (60), Expect = 7.0
Identities = 12/46 (26%), Positives = 23/46 (49%)
Frame = -3
Query: 266 FDANYENEDGGGENEAGPSTVTWEEPDFQSSLQMEQIREHIKNMFP 311
F + + G N+ GP T +++ + SSL +Q+ E ++ P
Sbjct: 303 FSSAPHTQTGQQSNQQGPDTFSYQFQEHHSSLPFQQLLEFFRDTSP 166
>TC223786 similar to UP|Q8I738 (Q8I738) TcC31.4, partial (11%)
Length = 649
Score = 27.7 bits (60), Expect = 7.0
Identities = 15/37 (40%), Positives = 20/37 (53%), Gaps = 3/37 (8%)
Frame = +3
Query: 159 IFYCLGKYYLVDAGYPTFM---GFLGPYKKNRYHLPQ 192
+FYCL Y+L P+F GFL ++K H PQ
Sbjct: 243 LFYCLFLYFL-----PSFSKESGFLHTHRKESLHFPQ 338
>TC219936 similar to UP|Q9LJL8 (Q9LJL8) Emb|CAB43653.1 (AT3g19120/MVI11_3),
partial (43%)
Length = 835
Score = 27.3 bits (59), Expect = 9.1
Identities = 13/41 (31%), Positives = 22/41 (52%)
Frame = +1
Query: 214 RNVIERSFGCCKAKWKILGSMPSYDLKTQNKIITACMALHN 254
R+V+ + K +WKIL + + ++ + I AC LHN
Sbjct: 292 RSVVVEAIALLKGRWKILQDL-NTGVRHAPQTIVACCVLHN 411
>TC216645 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [Contains:
Apg8FG], partial (76%)
Length = 640
Score = 27.3 bits (59), Expect = 9.1
Identities = 12/46 (26%), Positives = 23/46 (49%)
Frame = -1
Query: 266 FDANYENEDGGGENEAGPSTVTWEEPDFQSSLQMEQIREHIKNMFP 311
F + + G N+ GP T +++ + SSL +Q+ E ++ P
Sbjct: 346 FSSAPHTQTGQQSNQQGPDTFSYQFQEHHSSLPFQQLLEFFQDTSP 209
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.342 0.152 0.496
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,987,236
Number of Sequences: 63676
Number of extensions: 229581
Number of successful extensions: 1864
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1849
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1857
length of query: 312
length of database: 12,639,632
effective HSP length: 97
effective length of query: 215
effective length of database: 6,463,060
effective search space: 1389557900
effective search space used: 1389557900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0262.8


