

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0262.4
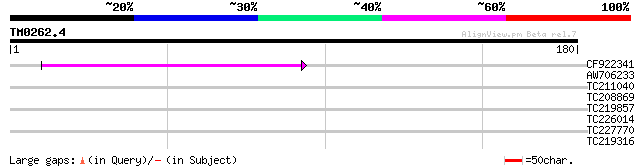
(180 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922341 46 1e-05
AW706233 29 1.4
TC211040 27 4.0
TC208869 similar to UP|Q7XJ60 (Q7XJ60) At3g47690, partial (54%) 27 4.0
TC219857 similar to UP|Q9LVY7 (Q9LVY7) Cytochrome P450-like, par... 27 5.2
TC226014 similar to UP|O23954 (O23954) Endo-1,4-beta-glucanase (... 27 5.2
TC227770 similar to UP|O04794 (O04794) Acyl-ACP thioesterase, pa... 26 8.9
TC219316 26 8.9
>CF922341
Length = 675
Score = 45.8 bits (107), Expect = 1e-05
Identities = 23/84 (27%), Positives = 37/84 (43%)
Frame = +1
Query: 11 VVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKMVISAASDAVKCRMFPSMFKGTAMA 70
+V + P K L + Y G T PK+HL + KM A + + F G A+
Sbjct: 385 LVPNIITPPKFKVLDFDKYKGTTCPKNHLKMYCQKMGAYAKDEELLIHSFQESLTGVAVT 564
Query: 71 WFTTLPLGSIVKFRDFSSKFLIQF 94
W+T L + ++D F+ Q+
Sbjct: 565 WYTNLEPSRVHSWKDLMVAFVRQY 636
>AW706233
Length = 376
Score = 28.9 bits (63), Expect = 1.4
Identities = 13/36 (36%), Positives = 18/36 (49%)
Frame = -3
Query: 11 VVREVAIPDNMKSLALEAYSGGTDPKDHLLYFSMKM 46
+V + P K L + Y G T PK+HL + KM
Sbjct: 263 LVXNIISPPKFKVLNFDKYKGTTCPKNHLKMYCQKM 156
>TC211040
Length = 951
Score = 27.3 bits (59), Expect = 4.0
Identities = 14/44 (31%), Positives = 23/44 (51%)
Frame = +2
Query: 67 TAMAWFTTLPLGSIVKFRDFSSKFLIQFSASKIKQVTIDDLYNV 110
T +W TLP+ +++ R K L+ S +K+ + LYNV
Sbjct: 737 TQGSWQNTLPVLTLLDLRHILFKSLVNNDKSGLKKQMMTKLYNV 868
>TC208869 similar to UP|Q7XJ60 (Q7XJ60) At3g47690, partial (54%)
Length = 982
Score = 27.3 bits (59), Expect = 4.0
Identities = 27/95 (28%), Positives = 42/95 (43%), Gaps = 9/95 (9%)
Frame = +2
Query: 40 LYFSMKMVISAASDAVKCRMFPSMFKGTAMAWFTTLPLGSIVKFRDFSSKFLIQ------ 93
L+ ++ + AAS AV+C+M F G +P+ V F + +IQ
Sbjct: 74 LHLNLSRIEEAASGAVQCQMMDMTFPG-------VVPMHK-VNFDAKTEYDMIQNYKILQ 229
Query: 94 --FSASKI-KQVTIDDLYNVRQLERETLKQYVKRY 125
F+ KI K V + L R L+ Q++KRY
Sbjct: 230 DVFNKLKIDKHVEVSRLVKGRPLDNLEFLQWLKRY 334
>TC219857 similar to UP|Q9LVY7 (Q9LVY7) Cytochrome P450-like, partial (36%)
Length = 1146
Score = 26.9 bits (58), Expect = 5.2
Identities = 21/84 (25%), Positives = 39/84 (46%), Gaps = 4/84 (4%)
Frame = +2
Query: 29 YSGGTDPKDHLLYFSMKMVISAASDAVKCRMF-PSMFKGTAMAWFTTLPLG---SIVKFR 84
+ G + PK LY+S + F PS F+GT A +T +P G S+ +
Sbjct: 509 FDGFSIPKGWKLYWSANSTHKNPEYFPEPEKFDPSRFEGTGPAPYTYVPFGGGPSMCPGK 688
Query: 85 DFSSKFLIQFSASKIKQVTIDDLY 108
+++ L+ F + +K+ + L+
Sbjct: 689 EYARMELLVFMHNLVKRFKCETLF 760
>TC226014 similar to UP|O23954 (O23954) Endo-1,4-beta-glucanase (Fragment) ,
partial (41%)
Length = 917
Score = 26.9 bits (58), Expect = 5.2
Identities = 12/35 (34%), Positives = 19/35 (54%)
Frame = +2
Query: 36 KDHLLYFSMKMVISAASDAVKCRMFPSMFKGTAMA 70
K HLLYFS + + + D V F ++++ T A
Sbjct: 761 KSHLLYFSTALDVFSVFDVVSFAAFCNLYRETVCA 865
>TC227770 similar to UP|O04794 (O04794) Acyl-ACP thioesterase, partial (56%)
Length = 1174
Score = 26.2 bits (56), Expect = 8.9
Identities = 10/26 (38%), Positives = 18/26 (68%)
Frame = +3
Query: 155 NKLSRKLARSMTEVRAQANTYILDEE 180
NKL+R+L++ EVR + +Y +D +
Sbjct: 225 NKLTRRLSKIPEEVRQEIGSYFVDSD 302
>TC219316
Length = 725
Score = 26.2 bits (56), Expect = 8.9
Identities = 13/30 (43%), Positives = 18/30 (59%)
Frame = -2
Query: 142 RAFKDGLLPGKLNNKLSRKLARSMTEVRAQ 171
RAF G +PG++ N + R+MTE R Q
Sbjct: 418 RAFPLGGVPGRIPNTTVLHINRAMTEFRRQ 329
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.135 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,646,180
Number of Sequences: 63676
Number of extensions: 57535
Number of successful extensions: 307
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 307
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 307
length of query: 180
length of database: 12,639,632
effective HSP length: 91
effective length of query: 89
effective length of database: 6,845,116
effective search space: 609215324
effective search space used: 609215324
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0262.4


