

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0253.6
(499 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
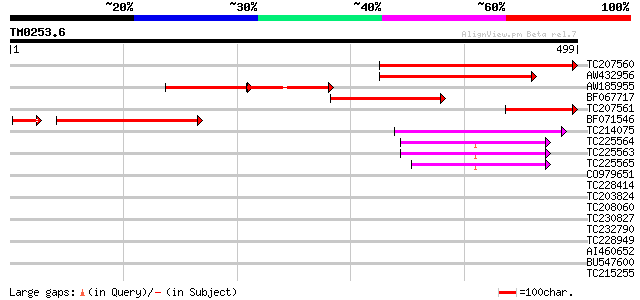
Score E
Sequences producing significant alignments: (bits) Value
TC207560 similar to UP|Q6GKU7 (Q6GKU7) At1g64880, partial (29%) 331 4e-91
AW432956 229 2e-60
AW185955 116 2e-54
BF067717 similar to GP|28200471|gb| endo-b1 4-mannanase 5B {Cell... 170 1e-42
TC207561 similar to UP|Q6GKU7 (Q6GKU7) At1g64880, partial (12%) 117 9e-27
BF071546 112 4e-26
TC214075 similar to UP|P93014 (P93014) 30S ribosomal protein S5,... 84 1e-16
TC225564 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 ... 44 1e-04
TC225563 similar to UP|RS2_ARATH (P49688) 40S ribosomal protein ... 44 1e-04
TC225565 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 ... 44 2e-04
CO979651 35 0.060
TC228414 weakly similar to UP|Q8LPR8 (Q8LPR8) AT5g22640/MDJ22_6,... 35 0.078
TC203824 similar to UP|Q9V3T8 (Q9V3T8) SR family splicing factor... 35 0.10
TC208060 weakly similar to UP|Q840B9 (Q840B9) Endo-b1,4-mannanas... 34 0.13
TC230827 weakly similar to GB|AAO42927.1|28416793|BT004681 At5g2... 33 0.39
TC232790 similar to UP|Q6K618 (Q6K618) Meprin and TRAF homology ... 32 0.50
TC228949 similar to UP|Q945N1 (Q945N1) AT5g50310/MXI22_1, partia... 32 0.86
AI460652 32 0.86
BU547600 homologue to GP|21554414|gb| glycoprotein homolog {Arab... 31 1.5
TC215255 similar to UP|Q39087 (Q39087) DNA-binding protein, part... 31 1.5
>TC207560 similar to UP|Q6GKU7 (Q6GKU7) At1g64880, partial (29%)
Length = 743
Score = 331 bits (849), Expect = 4e-91
Identities = 157/174 (90%), Positives = 169/174 (96%)
Frame = +2
Query: 326 RNELTEKKRRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGP 385
RNELTE+KR+PLYRKGFD +LI++NRTCKVTKGGQVVKYTAM+ACGNYNGV+GFAKAKGP
Sbjct: 8 RNELTEQKRKPLYRKGFDTRLIDMNRTCKVTKGGQVVKYTAMVACGNYNGVIGFAKAKGP 187
Query: 386 AVPVALQKAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQT 445
AVPVALQKAYEKCFQNLH+VERHEEHTIAHA+QTS+KKTKVYLWPAPT TGMKAGR V+
Sbjct: 188 AVPVALQKAYEKCFQNLHYVERHEEHTIAHAVQTSYKKTKVYLWPAPTTTGMKAGRSVEA 367
Query: 446 ILHLAGFKNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEKYLL 499
ILHLAG KNVKSKV+GSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEKYLL
Sbjct: 368 ILHLAGLKNVKSKVIGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEKYLL 529
>AW432956
Length = 430
Score = 229 bits (585), Expect = 2e-60
Identities = 108/138 (78%), Positives = 121/138 (87%)
Frame = +2
Query: 326 RNELTEKKRRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGP 385
RNELT++ R+PLYR G D LI++NRTC V+KGGQVVKYTAM+ACGNYNGV+G AKAKGP
Sbjct: 2 RNELTDQMRKPLYRTGSDTTLIDMNRTC*VSKGGQVVKYTAMVACGNYNGVIGCAKAKGP 181
Query: 386 AVPVALQKAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQT 445
AVPVAL KAYEKC QN+H+VERHEEHTIAHA+ TS+KKT VYLWPAPT TGMKAGR V
Sbjct: 182 AVPVALHKAYEKCLQNVHYVERHEEHTIAHAVHTSYKKTNVYLWPAPTTTGMKAGRSVDA 361
Query: 446 ILHLAGFKNVKSKVVGSR 463
ILHLAG KNVKSKV+ SR
Sbjct: 362 ILHLAGLKNVKSKVIDSR 415
>AW185955
Length = 443
Score = 116 bits (291), Expect(2) = 2e-54
Identities = 57/77 (74%), Positives = 65/77 (84%)
Frame = +2
Query: 209 LQNKLNRALRLVQWKEAFDPDNPDNYGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAA 268
LQNKLNRALRLV+WKEA+DPDNP NYGVIQ EQ PT+DA + A FEKEKQ +GD A
Sbjct: 218 LQNKLNRALRLVRWKEAYDPDNPANYGVIQREQ--PTADAKEEAEFEKEKQKQIEEGDNA 391
Query: 269 DDDDEQEFDDMKEKDNI 285
DD++EQEFDDMKEKDN+
Sbjct: 392 DDEEEQEFDDMKEKDNV 442
Score = 114 bits (286), Expect(2) = 2e-54
Identities = 53/76 (69%), Positives = 61/76 (79%)
Frame = +3
Query: 138 KKFDDFERKFKRHQDLLKNFTDADTLDDAFKWMNRIDKFEDKHFRLRPEYRVIGELMNRL 197
KK DFE+K +H DLL FT A+TLDDAFKWM +ID FE+KHFRLRPEYRVIGELMNRL
Sbjct: 6 KKVSDFEKKKDKHMDLLDKFTSAETLDDAFKWMAKIDNFENKHFRLRPEYRVIGELMNRL 185
Query: 198 KVVTDQKDKFILQNKL 213
KV T+ KDKF+ + L
Sbjct: 186 KVATELKDKFVCRTSL 233
>BF067717 similar to GP|28200471|gb| endo-b1 4-mannanase 5B {Cellvibrio
japonicus}, partial (4%)
Length = 307
Score = 170 bits (431), Expect = 1e-42
Identities = 81/101 (80%), Positives = 95/101 (93%)
Frame = +2
Query: 283 DNIILAKLDAIDKKLEEKLAELEYTFGKKGKVLEEEIRDLAEERNELTEKKRRPLYRKGF 342
DN+++AKL+AID+KLEEKLAELEYTFG+KGK LEEEI+DLAEERNELTE+KR+PLYRKGF
Sbjct: 2 DNVLMAKLEAIDRKLEEKLAELEYTFGRKGKALEEEIKDLAEERNELTEQKRKPLYRKGF 181
Query: 343 DVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAK 383
D +LI +NRTCKVT GGQVVKYTAM+A G+YNGV+GFAK K
Sbjct: 182 DTRLIXMNRTCKVT*GGQVVKYTAMVA*GDYNGVIGFAKPK 304
>TC207561 similar to UP|Q6GKU7 (Q6GKU7) At1g64880, partial (12%)
Length = 581
Score = 117 bits (294), Expect = 9e-27
Identities = 58/63 (92%), Positives = 60/63 (95%)
Frame = -1
Query: 437 MKAGRIVQTILHLAGFKNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEK 496
MKAGR V+ ILHLAG KNVKSKV+GSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEK
Sbjct: 581 MKAGRSVEAILHLAGLKNVKSKVIGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEK 402
Query: 497 YLL 499
YLL
Sbjct: 401 YLL 393
>BF071546
Length = 529
Score = 112 bits (281), Expect(2) = 4e-26
Identities = 61/129 (47%), Positives = 82/129 (63%), Gaps = 1/129 (0%)
Frame = +2
Query: 42 SSSYSGKSLYSTSSDARIVQDLLAQVEKDRLREKNERMRAGLDTADIDAEGEEDYMGVGP 101
SS G ++ S+ + + ++ A+VEKDR RE+NER+R GLDTADIDA+ E+DYMGVGP
Sbjct: 143 SSPILGNNVVFFSTGDKFLHEITAEVEKDRRRERNERLRLGLDTADIDADSEQDYMGVGP 322
Query: 102 LIEKLEKEKLKECPELMRYEEPTDSDSDEDEYEASQKKFDD-FERKFKRHQDLLKNFTDA 160
LI KL+ E L E EL RYEEPTDS+ D D +++ + F ++H DLL F
Sbjct: 323 LIAKLQNEMLMESAELNRYEEPTDSEDDSDVKP*RRRQEGE*FAEDERQHMDLLDIFYLG 502
Query: 161 DTLDDAFKW 169
+D A W
Sbjct: 503 VIVDHAL*W 529
Score = 23.9 bits (50), Expect(2) = 4e-26
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = +3
Query: 3 GYLQYGRQTFRQILRCSTSAGNVVRN 28
GY+ +GR R + R T+ G VV+N
Sbjct: 27 GYIHHGRTGLRHLFRFRTATG-VVKN 101
>TC214075 similar to UP|P93014 (P93014) 30S ribosomal protein S5, partial
(76%)
Length = 1140
Score = 84.3 bits (207), Expect = 1e-16
Identities = 49/152 (32%), Positives = 77/152 (50%)
Frame = +2
Query: 339 RKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKC 398
R GF+ +++ V R KV KGG+ +++ A++ G+ G VG K V A+QK+
Sbjct: 467 RDGFEERVVQVRRVTKVVKGGKQLRFRAVVVVGDKKGQVGVGVGKAKEVIAAVQKSAVNA 646
Query: 399 FQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSK 458
+N+ V + T H + KV L PA TG+ AG V+ +L +AG N K
Sbjct: 647 RRNIIKVPMTKYSTFPHRADGDYGAAKVMLRPASPGTGVIAGGSVRIVLEMAGVDNALGK 826
Query: 459 VVGSRNPHNTVKAVFKALNAIETPRDVQEKFG 490
+GS N N +A A+ ++ R+V E+ G
Sbjct: 827 QLGSNNALNNARATVAAVQKMKQFREVAEERG 922
>TC225564 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 homolog
(At3g57490), partial (93%)
Length = 1047
Score = 44.3 bits (103), Expect = 1e-04
Identities = 36/140 (25%), Positives = 61/140 (42%), Gaps = 8/140 (5%)
Frame = +1
Query: 345 KLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHF 404
+++ + K T+ GQ ++ A + G+ NG VG V A++ A ++
Sbjct: 334 EVMKITPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIP 513
Query: 405 VERH-------EEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKS 457
V R + HT+ + V + PAP +G+ A R+ + +L AG +V +
Sbjct: 514 VRRGYWGNKIGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFT 693
Query: 458 KVVGS-RNPHNTVKAVFKAL 476
GS + N VKA F L
Sbjct: 694 SSRGSTKTLGNFVKATFDCL 753
>TC225563 similar to UP|RS2_ARATH (P49688) 40S ribosomal protein S2, partial
(90%)
Length = 1117
Score = 44.3 bits (103), Expect = 1e-04
Identities = 36/140 (25%), Positives = 61/140 (42%), Gaps = 8/140 (5%)
Frame = +2
Query: 345 KLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHF 404
+++ + K T+ GQ ++ A + G+ NG VG V A++ A ++
Sbjct: 344 EVMKITPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIP 523
Query: 405 VERH-------EEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKS 457
V R + HT+ + V + PAP +G+ A R+ + +L AG +V +
Sbjct: 524 VRRGYWGNKIGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFT 703
Query: 458 KVVGS-RNPHNTVKAVFKAL 476
GS + N VKA F L
Sbjct: 704 SSRGSTKTLGNFVKATFDCL 763
>TC225565 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 homolog
(At3g57490), partial (92%)
Length = 1164
Score = 43.9 bits (102), Expect = 2e-04
Identities = 36/131 (27%), Positives = 57/131 (43%), Gaps = 8/131 (6%)
Frame = +1
Query: 354 KVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHFVERH----- 408
K T+ GQ ++ A + G+ NG VG V A++ A ++ V R
Sbjct: 367 KQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNK 546
Query: 409 --EEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGS-RNP 465
+ HT+ + V + PAP +G+ A R+ + +L AG +V + GS +
Sbjct: 547 IGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTL 726
Query: 466 HNTVKAVFKAL 476
N VKA F L
Sbjct: 727 GNFVKATFDCL 759
>CO979651
Length = 789
Score = 35.4 bits (80), Expect = 0.060
Identities = 36/147 (24%), Positives = 61/147 (41%), Gaps = 2/147 (1%)
Frame = -1
Query: 192 ELMNRLKVVTDQKDKFILQNKLNRALRLVQWKEAFDPDNPDNYGVIQHEQLGPTSDALQN 251
EL + K D++ +F+L + L + DP +H + D
Sbjct: 774 ELDEKKKKALDKQLEFLLGQTERYSTMLAE--NLGDPYKSAENNSAEHRKSIHCKDVHDV 601
Query: 252 AGFEKEKQTIQGQGDAADDDDEQEF--DDMKEKDNIILAKLDAIDKKLEEKLAELEYTFG 309
KE ++ Q DAAD+D+E + DD E D + + +A+ K EE+ EL
Sbjct: 600 INEPKEADVVEYQSDAADNDEEYDVQSDDELEDDERTIEQDEALITK-EERQEELAALRD 424
Query: 310 KKGKVLEEEIRDLAEERNELTEKKRRP 336
+ ++E ++ A E+ E K P
Sbjct: 423 EMDLPIQELLKRYAGEKGESVMKGSSP 343
>TC228414 weakly similar to UP|Q8LPR8 (Q8LPR8) AT5g22640/MDJ22_6, partial
(6%)
Length = 889
Score = 35.0 bits (79), Expect = 0.078
Identities = 25/78 (32%), Positives = 38/78 (48%), Gaps = 3/78 (3%)
Frame = +1
Query: 71 RLREKNERMRAG---LDTADIDAEGEEDYMGVGPLIEKLEKEKLKECPELMRYEEPTDSD 127
R+REK E +A LD D D EG+ D M ++ K+ E P + E+ D D
Sbjct: 118 RIREKEEEKKAKMGLLDEEDDDNEGDGDDMTSVTKQDEKAPAKVVEVPAEVEEEDDDDWD 297
Query: 128 SDEDEYEASQKKFDDFER 145
+ED+ ++Q F E+
Sbjct: 298 DEEDD-NSAQSSFGSVEQ 348
Score = 28.5 bits (62), Expect = 7.3
Identities = 17/49 (34%), Positives = 28/49 (56%), Gaps = 3/49 (6%)
Frame = +1
Query: 255 EKEKQTIQGQGDAADDDDEQEFDDM---KEKDNIILAKLDAIDKKLEEK 300
E+EK+ G D DDD+E + DDM ++D AK+ + ++EE+
Sbjct: 133 EEEKKAKMGLLDEEDDDNEGDGDDMTSVTKQDEKAPAKVVEVPAEVEEE 279
>TC203824 similar to UP|Q9V3T8 (Q9V3T8) SR family splicing factor SC35
(CG5442-PB) (LD32469p), partial (6%)
Length = 1532
Score = 34.7 bits (78), Expect = 0.10
Identities = 24/85 (28%), Positives = 42/85 (49%), Gaps = 5/85 (5%)
Frame = +2
Query: 40 ALSSSYSGKSLYSTSSDARI--VQDLLAQVEKDRLREKNERMRAGLDTADIDAEGEEDYM 97
+L + +G SD R+ ++ + +E + E+N+R+R +T D E E +
Sbjct: 623 SLENELAGLKKVKAESDVRVRDLERKIGVLETKEIEERNKRIRFEEETRDKIDEKEREIK 802
Query: 98 GVGPLIEKLEK---EKLKECPELMR 119
G +E+LEK EK E EL++
Sbjct: 803 GYRQKVEELEKVAAEKKTELDELVK 877
>TC208060 weakly similar to UP|Q840B9 (Q840B9) Endo-b1,4-mannanase 26B, partial
(5%)
Length = 2230
Score = 34.3 bits (77), Expect = 0.13
Identities = 16/64 (25%), Positives = 32/64 (50%)
Frame = +1
Query: 230 NPDNYGVIQHEQLGPTSDALQNAGFEKEKQTIQGQGDAADDDDEQEFDDMKEKDNIILAK 289
+P ++ ++ G ++ QN E + + +++ +EFDDMKE D L++
Sbjct: 1042 SPTGEADLEVDRHGEVANEDQNEVLETALHSEESMSQVTEENSNEEFDDMKEIDEEFLSE 1221
Query: 290 LDAI 293
LD +
Sbjct: 1222 LDTV 1233
>TC230827 weakly similar to GB|AAO42927.1|28416793|BT004681 At5g24660
{Arabidopsis thaliana;} , partial (41%)
Length = 570
Score = 32.7 bits (73), Expect = 0.39
Identities = 22/77 (28%), Positives = 37/77 (47%), Gaps = 2/77 (2%)
Frame = -3
Query: 295 KKLEEKLAELEYTFGKKGKVLEEEIR--DLAEERNELTEKKRRPLYRKGFDVKLINVNRT 352
++ +EK +L G V EE +R L +ERN+ + R L R GF+V +
Sbjct: 403 ERRKEKKIDLRARMKGNGGVEEERVRLRKLVDERNDASVVLTRLLQRLGFEVTELGAEAL 224
Query: 353 CKVTKGGQVVKYTAMLA 369
+ + G+ ++ A LA
Sbjct: 223 LRFLRHGEAIQCGAELA 173
>TC232790 similar to UP|Q6K618 (Q6K618) Meprin and TRAF homology
domain-containing protein-like, partial (67%)
Length = 780
Score = 32.3 bits (72), Expect = 0.50
Identities = 20/75 (26%), Positives = 39/75 (51%), Gaps = 1/75 (1%)
Frame = +3
Query: 273 EQEFDDMKEKDNIILAKLDAIDKKLEEKLAELEYTFGKKGKVLEEEIRDLAEERNELTEK 332
E + + ++ + + +AKL A K L ++L + E + +E++ + +E+N L E+
Sbjct: 18 ESQLEWIRSERDDEIAKLSAEKKALHDRLHDAETQLSQLKSRKRDELKKVVKEKNALAER 197
Query: 333 -KRRPLYRKGFDVKL 346
K RK FD +L
Sbjct: 198 LKNAEAARKRFDEEL 242
>TC228949 similar to UP|Q945N1 (Q945N1) AT5g50310/MXI22_1, partial (27%)
Length = 1134
Score = 31.6 bits (70), Expect = 0.86
Identities = 16/38 (42%), Positives = 23/38 (60%)
Frame = +3
Query: 266 DAADDDDEQEFDDMKEKDNIILAKLDAIDKKLEEKLAE 303
+A+DDD+E E DD E D L D D++ EE+ A+
Sbjct: 159 EASDDDEENEDDDEDESDGDSLTDEDEDDEEEEEEEAQ 272
>AI460652
Length = 394
Score = 31.6 bits (70), Expect = 0.86
Identities = 16/43 (37%), Positives = 25/43 (57%), Gaps = 4/43 (9%)
Frame = +3
Query: 108 KEKLKECPELMRYEEPTDSDSDEDE----YEASQKKFDDFERK 146
K+K+ YE+ +D+D D+DE ++ S +KFDDF K
Sbjct: 165 KKKIIPVASRPYYEDESDNDHDDDEGKHPHQTSPRKFDDFLTK 293
>BU547600 homologue to GP|21554414|gb| glycoprotein homolog {Arabidopsis
thaliana}, partial (6%)
Length = 659
Score = 30.8 bits (68), Expect = 1.5
Identities = 22/75 (29%), Positives = 36/75 (47%)
Frame = -2
Query: 74 EKNERMRAGLDTADIDAEGEEDYMGVGPLIEKLEKEKLKECPELMRYEEPTDSDSDEDEY 133
EK + D D D E E++ +G G +I + + + P+++ E S +D DE
Sbjct: 427 EKESFLVESNDDEDEDTETEDEDVGGGRIIVQNQNNDSGKGPQVIGGES---SKTDGDEG 257
Query: 134 EASQKKFDDFERKFK 148
KK D+F KF+
Sbjct: 256 PDVDKKADEFIAKFR 212
>TC215255 similar to UP|Q39087 (Q39087) DNA-binding protein, partial (67%)
Length = 732
Score = 30.8 bits (68), Expect = 1.5
Identities = 25/89 (28%), Positives = 43/89 (48%), Gaps = 6/89 (6%)
Frame = +2
Query: 252 AGFEKEKQTI------QGQGDAADDDDEQEFDDMKEKDNIILAKLDAIDKKLEEKLAELE 305
A EKEK+ + + A++ ++ + +N A L+A KKLEE+L + +
Sbjct: 248 AEVEKEKKLSYVKAWEESEKAKAENRAQKHLSAIAAWENSKKAALEAELKKLEEQLEKKK 427
Query: 306 YTFGKKGKVLEEEIRDLAEERNELTEKKR 334
+G+K K + AEE+ + E KR
Sbjct: 428 AEYGEKMKNKVALVHKEAEEKRAMIEAKR 514
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,936,570
Number of Sequences: 63676
Number of extensions: 192309
Number of successful extensions: 1228
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 1184
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1223
length of query: 499
length of database: 12,639,632
effective HSP length: 101
effective length of query: 398
effective length of database: 6,208,356
effective search space: 2470925688
effective search space used: 2470925688
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0253.6


