

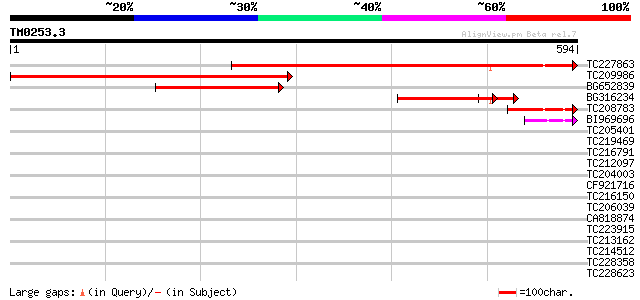
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0253.3
(594 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227863 572 e-163
TC209986 weakly similar to UP|Q811Z9 (Q811Z9) N-terminal aceyltr... 543 e-155
BG652839 similar to GP|13365578|db P0686E09.16 {Oryza sativa (ja... 256 2e-68
BG316234 homologue to GP|1653666|dbj| chloride channel protein {... 144 1e-34
TC208783 99 5e-21
BI969696 42 8e-04
TC205401 similar to GB|AAP37850.1|30725656|BT008491 At4g26630 {A... 39 0.008
TC219469 similar to UP|Q9ZS03 (Q9ZS03) RNA helicase (Fragment), ... 37 0.025
TC216791 similar to UP|O23144 (O23144) Proton pump interactor (A... 36 0.042
TC212097 similar to UP|Q84MB7 (Q84MB7) At3g62970, partial (80%) 35 0.072
TC204003 34 0.16
CF921716 34 0.21
TC216150 weakly similar to UP|MNN4_YEAST (P36044) MNN4 protein, ... 34 0.21
TC206039 33 0.36
CA818874 33 0.36
TC223915 similar to UP|Q7PZD4 (Q7PZD4) AgCP9387 (Fragment), part... 33 0.47
TC213162 32 0.80
TC214512 homologue to UP|O65334 (O65334) SAR DNA-binding protein... 32 0.80
TC228358 weakly similar to GB|AAH51484.1|30185799|BC051484 GCIP-... 32 1.0
TC228623 similar to UP|Q9SA21 (Q9SA21) F3O9.2 protein, partial (... 32 1.0
>TC227863
Length = 1671
Score = 572 bits (1473), Expect = e-163
Identities = 290/367 (79%), Positives = 316/367 (86%), Gaps = 5/367 (1%)
Frame = +3
Query: 233 LRKMTLRTYVEMLKFQDRLHSHVYFHKAAVGAIRCYIKLHDSPPKSTAEEDDDMSKLLPS 292
LRKMT RTYVEMLKFQ +HSH YFHKAA GAIRCYIKLHDSPPKSTAEEDD+MSKLLPS
Sbjct: 9 LRKMTXRTYVEMLKFQXGVHSHAYFHKAAAGAIRCYIKLHDSPPKSTAEEDDNMSKLLPS 188
Query: 293 QKKKLRQKQRKAEARAKKEAEEKNEESSASGISKSGKRQAKPVDPDPRGEKLLQVEDPLL 352
QKKK+RQKQRKAEARAKKEAEEKNEESSASG+SKSGKR KPVDPDP GEKLLQVEDPL
Sbjct: 189 QKKKMRQKQRKAEARAKKEAEEKNEESSASGVSKSGKRHVKPVDPDPNGEKLLQVEDPLS 368
Query: 353 EATKYLKLLQKNSPDSLETHLLSFELHMRRQKILLAFQAVKQLLRLDAEHPDSHRCLIKF 412
EATKYLKLLQKNSPDSLETHLLSFEL+ R+QKILLA QAVKQLLRLDAEHPDSHRCLIKF
Sbjct: 369 EATKYLKLLQKNSPDSLETHLLSFELYTRKQKILLALQAVKQLLRLDAEHPDSHRCLIKF 548
Query: 413 FHKVGSLNTPVNDSEKLVWSVVEAERQTISQLQGKSLFETNKSFLEKYEDSLMHRAAFGE 472
FHKVGS+N PV DSEKL+WSV+EAER TISQL KSLFE N SFLEK++DSLMHRAAF E
Sbjct: 549 FHKVGSMNAPVTDSEKLIWSVLEAERPTISQLHEKSLFEANNSFLEKHKDSLMHRAAFAE 728
Query: 473 MMYVLDPSRRSEAVKIIEGSTNSPVSRNGT-----NWKLEDCIEVHKLLGTVLVDKDAAL 527
++++LD +R+SEAVK +E STN+ V RNG W L DCI VHKLL TVL D+DA L
Sbjct: 729 ILHILDSNRKSEAVKFVEDSTNNIVPRNGALGPIREWNLTDCIAVHKLLETVLADQDAGL 908
Query: 528 RWKVRCAEIFPYSVYFEGRHSSASPNSALNQICKKSFENGSSSHSVGNCKVESITSNGKL 587
RWKVRCAE FPYS YFEG HSSASPNSA +Q+ +K+ EN S +HSV V SITSNGKL
Sbjct: 909 RWKVRCAEYFPYSTYFEGCHSSASPNSAFSQL-RKNSENESLNHSVDGQNVGSITSNGKL 1085
Query: 588 DTFKDLT 594
+ FKDLT
Sbjct: 1086EAFKDLT 1106
>TC209986 weakly similar to UP|Q811Z9 (Q811Z9) N-terminal aceyltransferase 1,
partial (18%)
Length = 1375
Score = 543 bits (1400), Expect = e-155
Identities = 262/296 (88%), Positives = 284/296 (95%)
Frame = +3
Query: 1 RIPLDFLQGDRFREAADSYMRPLLTKGVPSLFSDLSSLYDHPGKANILEQLILELENSIR 60
RIPLDFLQGD+F+EAA++Y+RPLLTKG+PSLFSDLSSLY+ PGKA+ILEQ+ILE+E+SI+
Sbjct: 486 RIPLDFLQGDKFQEAANNYIRPLLTKGIPSLFSDLSSLYNQPGKADILEQIILEIESSIK 665
Query: 61 TTGTYPGRVEKEPPSTLMWILFLLSQHYDRRSQYEIALSKINEAIEHTPTVIDLYSVKSR 120
TT YPG +EKEPPSTLMW LFLL+QHYDRR QYEIAL KINEAI+HTPTVIDLYSVKSR
Sbjct: 666 TTSQYPGGMEKEPPSTLMWTLFLLAQHYDRRGQYEIALFKINEAIDHTPTVIDLYSVKSR 845
Query: 121 ILKHAGDLAAAAAFADEARCMDLSDRYVNSECVKRMLQADQVVLAEKTAVLFTKEGDQHN 180
ILKHAGDL AAAAFADEARCMDL+DRYVNSECVKRMLQADQV LAEKTAVLFTK+GDQHN
Sbjct: 846 ILKHAGDLVAAAAFADEARCMDLADRYVNSECVKRMLQADQVALAEKTAVLFTKDGDQHN 1025
Query: 181 NLHDMQCMWYELASAESYFRQGDLGLALKKFLAVEKHYTDITEDQFDFHSYCLRKMTLRT 240
NLHDMQCMWYELASAES+FRQG+LG+ALKKFLAVEKHY DITEDQFDFHSYCLRKMTLRT
Sbjct: 1026NLHDMQCMWYELASAESHFRQGNLGMALKKFLAVEKHYADITEDQFDFHSYCLRKMTLRT 1205
Query: 241 YVEMLKFQDRLHSHVYFHKAAVGAIRCYIKLHDSPPKSTAEEDDDMSKLLPSQKKK 296
YVEMLKFQD+LHSH YFHKAA GAIRCYIKLHDSPPKSTAEED+DMSKLLPSQKKK
Sbjct: 1206YVEMLKFQDQLHSHAYFHKAAAGAIRCYIKLHDSPPKSTAEEDNDMSKLLPSQKKK 1373
>BG652839 similar to GP|13365578|db P0686E09.16 {Oryza sativa (japonica
cultivar-group)}, partial (19%)
Length = 404
Score = 256 bits (654), Expect = 2e-68
Identities = 123/134 (91%), Positives = 126/134 (93%)
Frame = +2
Query: 153 VKRMLQADQVVLAEKTAVLFTKEGDQHNNLHDMQCMWYELASAESYFRQGDLGLALKKFL 212
VKRMLQADQV LAEKTAVLFTK+GDQHNNLHDMQCMWYELAS ESYFRQGDLG ALKKFL
Sbjct: 2 VKRMLQADQVALAEKTAVLFTKDGDQHNNLHDMQCMWYELASGESYFRQGDLGRALKKFL 181
Query: 213 AVEKHYTDITEDQFDFHSYCLRKMTLRTYVEMLKFQDRLHSHVYFHKAAVGAIRCYIKLH 272
AVEKHY DITEDQFDFHSYCLRKMTL TYVEMLKFQD+LHSH YFHKAA GAIR YIKLH
Sbjct: 182 AVEKHYADITEDQFDFHSYCLRKMTLCTYVEMLKFQDQLHSHAYFHKAAAGAIRGYIKLH 361
Query: 273 DSPPKSTAEEDDDM 286
DSPPKSTAEEDD+M
Sbjct: 362 DSPPKSTAEEDDNM 403
>BG316234 homologue to GP|1653666|dbj| chloride channel protein
{Synechocystis sp. PCC 6803}, partial (1%)
Length = 421
Score = 144 bits (363), Expect = 1e-34
Identities = 73/106 (68%), Positives = 88/106 (82%), Gaps = 1/106 (0%)
Frame = +2
Query: 407 RCLIKFFHKVGSLNTPVNDSEKLVWSVVEAERQTISQLQGKSLFETNKSFLEKYEDSLMH 466
R LIKFF+KVGS+ PV DSEKL+WSV+EAERQTISQL GKSLFETN SFLEK+EDSL H
Sbjct: 2 RGLIKFFNKVGSMIAPVTDSEKLIWSVLEAERQTISQLHGKSLFETNNSFLEKHEDSLTH 181
Query: 467 RAAFGEMMYVLDPSRRSEAVKIIEGSTNSPVSRNGTNWKLE-DCIE 511
RAAFGE +Y+LDP+RRSEAVK+IEGS N+ V + +E +C++
Sbjct: 182 RAAFGETLYILDPNRRSEAVKLIEGSPNNIVPTHAIFCLIEMECLD 319
Score = 62.8 bits (151), Expect = 4e-10
Identities = 31/47 (65%), Positives = 34/47 (71%), Gaps = 5/47 (10%)
Frame = +1
Query: 492 STNSPVSRNGT-----NWKLEDCIEVHKLLGTVLVDKDAALRWKVRC 533
S N +RNG WKL DC+ VHKLLGTVLVD+DAALRWKVRC
Sbjct: 280 SCNILFNRNGVLGPIREWKLIDCVAVHKLLGTVLVDQDAALRWKVRC 420
>TC208783
Length = 769
Score = 99.0 bits (245), Expect = 5e-21
Identities = 52/73 (71%), Positives = 59/73 (80%)
Frame = +2
Query: 522 DKDAALRWKVRCAEIFPYSVYFEGRHSSASPNSALNQICKKSFENGSSSHSVGNCKVESI 581
D+DAA RWK+RCAE+FPYS YFEG SSASPNSA NQI +KS E GSS+H VG+ ES
Sbjct: 2 DQDAASRWKMRCAELFPYSTYFEGICSSASPNSAFNQI-RKSTETGSSNHWVGDHNAES- 175
Query: 582 TSNGKLDTFKDLT 594
TSNGKL+ FKDLT
Sbjct: 176 TSNGKLEAFKDLT 214
>BI969696
Length = 725
Score = 42.0 bits (97), Expect = 8e-04
Identities = 27/55 (49%), Positives = 33/55 (59%)
Frame = -2
Query: 540 SVYFEGRHSSASPNSALNQICKKSFENGSSSHSVGNCKVESITSNGKLDTFKDLT 594
S EG S AS A Q+CK S E+GS + VG+ ES TSNG L+ FKDL+
Sbjct: 721 STXXEGXXSCASHICAXTQMCKSS-ESGSYTQWVGDHNAES-TSNGNLEAFKDLS 563
>TC205401 similar to GB|AAP37850.1|30725656|BT008491 At4g26630 {Arabidopsis
thaliana;} , partial (16%)
Length = 912
Score = 38.5 bits (88), Expect = 0.008
Identities = 36/144 (25%), Positives = 59/144 (40%), Gaps = 10/144 (6%)
Frame = +3
Query: 274 SPPK----------STAEEDDDMSKLLPSQKKKLRQKQRKAEARAKKEAEEKNEESSASG 323
SPPK S ++ED D S + S+KKK + ++ A K A E+ E G
Sbjct: 141 SPPKRAPKKPSSNLSKSDEDSDESPKVFSRKKKNEKGGKQKTATPTKSASEEKTEKVTRG 320
Query: 324 ISKSGKRQAKPVDPDPRGEKLLQVEDPLLEATKYLKLLQKNSPDSLETHLLSFELHMRRQ 383
K K +++P D Q+ D + E K + D L+ F++ + +
Sbjct: 321 KGKK-KEKSRPSDN--------QLRDAICEILKEVNFNTATFTDILKKLAKQFDMDLTPR 473
Query: 384 KILLAFQAVKQLLRLDAEHPDSHR 407
K + ++L +L E D R
Sbjct: 474 KASIKSMIQEELTKLADEADDEDR 545
>TC219469 similar to UP|Q9ZS03 (Q9ZS03) RNA helicase (Fragment), partial
(66%)
Length = 1062
Score = 37.0 bits (84), Expect = 0.025
Identities = 26/83 (31%), Positives = 41/83 (49%), Gaps = 6/83 (7%)
Frame = +3
Query: 294 KKKLRQKQRKAEARAKKEAEEKNEESSASGISKSGKRQAKPVD------PDPRGEKLLQV 347
+ K R+KQRK +AKKEA+EK + K K P D R ++ ++
Sbjct: 783 RDKSREKQRKKNLQAKKEAKEKEPKP-----QKPKKTPNAPTDMRKKTARQRRAQQTMED 947
Query: 348 EDPLLEATKYLKLLQKNSPDSLE 370
E+ L++ + LK L+K + D E
Sbjct: 948 EEELMQEYRLLKKLKKGTIDENE 1016
>TC216791 similar to UP|O23144 (O23144) Proton pump interactor
(AT4g27500/F27G19_100), partial (25%)
Length = 1385
Score = 36.2 bits (82), Expect = 0.042
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 1/71 (1%)
Frame = +3
Query: 281 EEDDDMSKLLPSQKKKLRQKQR-KAEARAKKEAEEKNEESSASGISKSGKRQAKPVDPDP 339
EE+ +K +KKKL +K KA RA+KEAE+K ++ KSG + +P
Sbjct: 657 EEEIAKAKQALERKKKLAEKAAAKAAIRAQKEAEKKLKDREKKAKKKSGTAAVANPEDEP 836
Query: 340 RGEKLLQVEDP 350
+ E +++V +P
Sbjct: 837 KDE-VVEVTEP 866
>TC212097 similar to UP|Q84MB7 (Q84MB7) At3g62970, partial (80%)
Length = 905
Score = 35.4 bits (80), Expect = 0.072
Identities = 21/59 (35%), Positives = 28/59 (46%), Gaps = 5/59 (8%)
Frame = +1
Query: 369 LETHLLSFELHMRRQKILLAFQAVKQLLRLDA-----EHPDSHRCLIKFFHKVGSLNTP 422
L+ HLLS H+R +L F + RLDA + H I+FFH V + TP
Sbjct: 49 LQKHLLSLPFHLRLSPSMLLFCVYSFVPRLDASTIRDDAKFEHHAAIRFFHVVTATTTP 225
>TC204003
Length = 809
Score = 34.3 bits (77), Expect = 0.16
Identities = 21/62 (33%), Positives = 28/62 (44%)
Frame = +2
Query: 295 KKLRQKQRKAEARAKKEAEEKNEESSASGISKSGKRQAKPVDPDPRGEKLLQVEDPLLEA 354
K+ +K+ + AKKE E+K E G + K K DP P V DP+LE
Sbjct: 131 KEPEKKEDPKKVEAKKEGEKKEEGKKEEGKKEEKKEDEKKKDPPP-------VPDPVLEW 289
Query: 355 TK 356
K
Sbjct: 290 VK 295
>CF921716
Length = 367
Score = 33.9 bits (76), Expect = 0.21
Identities = 19/57 (33%), Positives = 32/57 (55%), Gaps = 6/57 (10%)
Frame = +2
Query: 293 QKKKLRQKQR------KAEARAKKEAEEKNEESSASGISKSGKRQAKPVDPDPRGEK 343
QKKK ++K+R K + + KK+ +K ++ + GK++ +P P PRGEK
Sbjct: 59 QKKKTQKKKRGGPPTEKKKKKKKKKGGKKKKKEGGKNKTPPGKKKRRP--PTPRGEK 223
>TC216150 weakly similar to UP|MNN4_YEAST (P36044) MNN4 protein, partial (5%)
Length = 928
Score = 33.9 bits (76), Expect = 0.21
Identities = 31/135 (22%), Positives = 63/135 (45%), Gaps = 4/135 (2%)
Frame = +3
Query: 234 RKMTLRTYVEMLKFQDRLHSHVYFHKAAVGAIRCYIKLHDSPPKSTAEEDDDMSKLLPSQ 293
+K+T+ V+ + +L + +VG + K + PK + ++ K P +
Sbjct: 282 KKLTVIGTVDPVNVVSKLRKYWQTDIVSVGPAKEPEKKEE--PKKEEPKKEEEKKEEPKK 455
Query: 294 KKKLRQKQRKAEARAKKEAEEKNEESSASGISK--SGKRQAKPVDPDPRGE--KLLQVED 349
+ + ++++ K E K+ EEK EE+ K K++A P PDP E K + +
Sbjct: 456 EGEAKKEEEKKEE--PKKEEEKKEETKKEEEKKEEEKKKEAPPPPPDPVLELVKAYRAYN 629
Query: 350 PLLEATKYLKLLQKN 364
P + Y++ +++N
Sbjct: 630 PHMTTYYYVQSMEEN 674
>TC206039
Length = 834
Score = 33.1 bits (74), Expect = 0.36
Identities = 20/77 (25%), Positives = 35/77 (44%)
Frame = -1
Query: 267 CYIKLHDSPPKSTAEEDDDMSKLLPSQKKKLRQKQRKAEARAKKEAEEKNEESSASGISK 326
CYI K ++ K +KKK ++K++K + KK+ ++K G +K
Sbjct: 294 CYISALKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKN*KKKKKKKKKKKXXXGGGGGTK 115
Query: 327 SGKRQAKPVDPDPRGEK 343
+++ K P P G K
Sbjct: 114 KXQKEGKKGAP-PGGAK 67
>CA818874
Length = 444
Score = 33.1 bits (74), Expect = 0.36
Identities = 15/58 (25%), Positives = 33/58 (56%)
Frame = +3
Query: 293 QKKKLRQKQRKAEARAKKEAEEKNEESSASGISKSGKRQAKPVDPDPRGEKLLQVEDP 350
+KKK ++K++K + + KK+ ++K ++ K G + KP+ G+K+ ++ P
Sbjct: 171 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKGGAPKKKPILTPGVGKKIFFLKGP 344
>TC223915 similar to UP|Q7PZD4 (Q7PZD4) AgCP9387 (Fragment), partial (7%)
Length = 643
Score = 32.7 bits (73), Expect = 0.47
Identities = 15/40 (37%), Positives = 25/40 (62%)
Frame = +3
Query: 280 AEEDDDMSKLLPSQKKKLRQKQRKAEARAKKEAEEKNEES 319
A E+ K QKKKLR+K++K++ A ++ +EK + S
Sbjct: 270 AAEERTAKKRAKRQKKKLRKKEKKSKLNAGEQLQEKEDSS 389
>TC213162
Length = 426
Score = 32.0 bits (71), Expect = 0.80
Identities = 15/22 (68%), Positives = 17/22 (77%)
Frame = +2
Query: 573 VGNCKVESITSNGKLDTFKDLT 594
VG+ ES TSNGKL+ FKDLT
Sbjct: 2 VGDHNAESGTSNGKLEAFKDLT 67
>TC214512 homologue to UP|O65334 (O65334) SAR DNA-binding protein-1, partial
(7%)
Length = 1535
Score = 32.0 bits (71), Expect = 0.80
Identities = 18/72 (25%), Positives = 34/72 (47%)
Frame = +2
Query: 232 CLRKMTLRTYVEMLKFQDRLHSHVYFHKAAVGAIRCYIKLHDSPPKSTAEEDDDMSKLLP 291
C +K+ +TY+ M ++QD L HVY + + + K + +
Sbjct: 590 CSKKIPHKTYICMFRWQDTLDMHVYSKNTSQKYAHV*VAKYLMKKKKKKKTNKKKK*TND 769
Query: 292 SQKKKLRQKQRK 303
++KKK R+K++K
Sbjct: 770 NEKKKRRRKKKK 805
>TC228358 weakly similar to GB|AAH51484.1|30185799|BC051484 GCIP-interacting
protein p29 {Mus musculus;} , partial (28%)
Length = 1460
Score = 31.6 bits (70), Expect = 1.0
Identities = 23/60 (38%), Positives = 35/60 (58%), Gaps = 6/60 (10%)
Frame = +2
Query: 273 DSPPKSTAEEDDDMSKLLPSQKK--KLRQKQRKAEARAKKE--AEEKNEE--SSASGISK 326
D P AE++ D +KL QKK +LR K ++A+ R + E AE+K E + + G+SK
Sbjct: 371 DQPAVQEAEQEVDYTKLSARQKKWMELRSKMQEAKKRNQIEIAAEKKRMEAPTESRGVSK 550
>TC228623 similar to UP|Q9SA21 (Q9SA21) F3O9.2 protein, partial (85%)
Length = 1082
Score = 31.6 bits (70), Expect = 1.0
Identities = 21/63 (33%), Positives = 28/63 (44%)
Frame = +3
Query: 294 KKKLRQKQRKAEARAKKEAEEKNEESSASGISKSGKRQAKPVDPDPRGEKLLQVEDPLLE 353
+K+ RKAEAR E EEK E S + K R + PV E + E+ E
Sbjct: 279 EKRAEAAARKAEARRLAEQEEKELEKSMKKVDKKATRVSIPVPKVTEVELRRRREEEQAE 458
Query: 354 ATK 356
A +
Sbjct: 459 AER 467
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.132 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,629,801
Number of Sequences: 63676
Number of extensions: 329796
Number of successful extensions: 1715
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 1693
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1709
length of query: 594
length of database: 12,639,632
effective HSP length: 103
effective length of query: 491
effective length of database: 6,081,004
effective search space: 2985772964
effective search space used: 2985772964
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0253.3


