

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
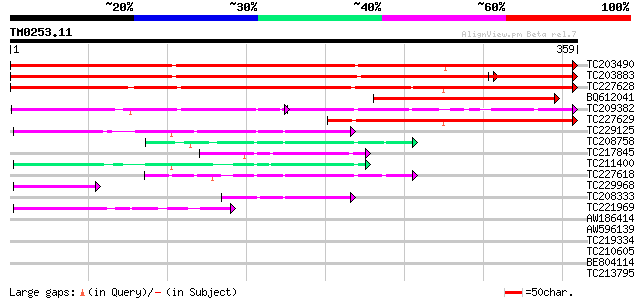
Query= TM0253.11
(359 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203490 weakly similar to GB|AAL15368.1|16323268|AY057738 At1g1... 582 e-167
TC203883 weakly similar to GB|AAL15368.1|16323268|AY057738 At1g1... 487 e-160
TC227628 similar to UP|Q6H8E0 (Q6H8E0) Kelch repeat-containing F... 451 e-127
BQ612041 198 3e-51
TC209382 125 3e-51
TC227629 similar to UP|Q6H8E0 (Q6H8E0) Kelch repeat-containing F... 189 2e-48
TC229125 weakly similar to GB|AAC13686.1|3047308|AF056929 sarcos... 69 3e-12
TC208758 similar to UP|Q8L736 (Q8L736) Predicted protein, partia... 59 3e-09
TC217845 similar to UP|Q6K649 (Q6K649) Kelch repeat-containing F... 57 1e-08
TC211400 similar to UP|Q9FKJ0 (Q9FKJ0) Emb|CAB55405.1, partial (... 55 5e-08
TC227618 similar to UP|Q9FKJ0 (Q9FKJ0) Emb|CAB55405.1, partial (... 53 2e-07
TC229968 similar to UP|O64797 (O64797) T1F15.5 protein, partial ... 52 5e-07
TC208333 weakly similar to GB|AAC13686.1|3047308|AF056929 sarcos... 47 1e-05
TC221969 45 5e-05
AW186414 37 0.011
AW596139 weakly similar to GP|18086559|gb| At2g24540/F25P17.16 {... 35 0.069
TC219334 weakly similar to UP|Q9FUZ9 (Q9FUZ9) SKP1 interacting p... 33 0.15
TC210605 weakly similar to UP|Q9FV01 (Q9FV01) SKP1 interacting p... 32 0.34
BE804114 32 0.58
TC213795 30 1.7
>TC203490 weakly similar to GB|AAL15368.1|16323268|AY057738 At1g15670/F7H2_1
{Arabidopsis thaliana;} , partial (38%)
Length = 1558
Score = 582 bits (1501), Expect = e-167
Identities = 281/364 (77%), Positives = 309/364 (84%), Gaps = 5/364 (1%)
Frame = +2
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 60
MELISGLPEDVARDCLIRV Y+QFPAV + CKGW+ EI SP+F RRRR T QKILVT
Sbjct: 158 MELISGLPEDVARDCLIRVPYDQFPAVASVCKGWSAEIHSPDFHRRRRTTKQAQKILVTV 337
Query: 61 QARFDSD-TCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVG 119
Q++ DSD T GLL K+TTNPVYRLSVFEP+TG+W EL + PE GLPMFCQIAGVG
Sbjct: 338 QSKIDSDKTRTGLLAKSTTNPVYRLSVFEPKTGSWCELPLGPEL--AFGLPMFCQIAGVG 511
Query: 120 YELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGH 179
++LVVMGGWDP+SWKASNSVFIYNFLSA WRRGADMPGGPRTFFAC SDQ+RTVYVAGGH
Sbjct: 512 FDLVVMGGWDPDSWKASNSVFIYNFLSAKWRRGADMPGGPRTFFACASDQNRTVYVAGGH 691
Query: 180 DEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFER 239
DEEKNAL+SALAYDVA D WVPLPDM+RERDECKAVFR G G L VVGGYCTEMQGRFER
Sbjct: 692 DEEKNALRSALAYDVAMDVWVPLPDMSRERDECKAVFRRG-GALCVVGGYCTEMQGRFER 868
Query: 240 SAEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGDA----MYMCRGGDVVALQGNTWQTVA 295
SAE FDV W+WGPVEEEFLD CPRTC+DG D M+MCRGGDVVAL G+TW+ VA
Sbjct: 869 SAEVFDVAKWKWGPVEEEFLDAAACPRTCVDGADGAEGRMFMCRGGDVVALHGDTWRNVA 1048
Query: 296 KVPSEIRNVACMGAWERSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQSYC 355
KVP EIRNVAC+G WE +L+IGSSGFGEPHMGF+LDLK+ W KL SP+DYTGHVQS C
Sbjct: 1049KVPGEIRNVACVGVWEGLMLVIGSSGFGEPHMGFVLDLKNRAWTKLASPEDYTGHVQSGC 1228
Query: 356 LLEI 359
LL+I
Sbjct: 1229LLQI 1240
>TC203883 weakly similar to GB|AAL15368.1|16323268|AY057738 At1g15670/F7H2_1
{Arabidopsis thaliana;} , partial (61%)
Length = 1668
Score = 487 bits (1253), Expect(2) = e-160
Identities = 240/312 (76%), Positives = 264/312 (83%), Gaps = 3/312 (0%)
Frame = +3
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 60
MELISGLPEDVARDCLIR+ YEQFPAV + CKGWNTEI SP+F RRRR T Q+ILVT
Sbjct: 207 MELISGLPEDVARDCLIRIPYEQFPAVASVCKGWNTEIHSPDFHRRRRTTKQAQEILVTV 386
Query: 61 QARFDSD-TCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVG 119
Q+ DS+ T GLL K+TTNPVYRLSVFEP+TG+WSEL + PE GLPMFC+IAGVG
Sbjct: 387 QSNIDSEKTRTGLLAKSTTNPVYRLSVFEPKTGSWSELPLGPEL--AFGLPMFCRIAGVG 560
Query: 120 YELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQ-DRTVYVAGG 178
++LVVMGGWDP+SWKASNSVFIYNFLSA WRRGADMPGGPRTFFAC SDQ ++TVYVAGG
Sbjct: 561 FDLVVMGGWDPDSWKASNSVFIYNFLSAKWRRGADMPGGPRTFFACASDQNNQTVYVAGG 740
Query: 179 HDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFE 238
HDEEKNAL+S LAYDVA D WVPLPDM+RERDECKAVFR GA L VVGGYCTEMQGRFE
Sbjct: 741 HDEEKNALRSVLAYDVARDLWVPLPDMSRERDECKAVFRRGA--LCVVGGYCTEMQGRFE 914
Query: 239 RSAEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGD-AMYMCRGGDVVALQGNTWQTVAKV 297
RSAE FDV TW+WGPVEEEF+D CPRT +DGGD AMYMC GGDVVALQG+TW+ VAKV
Sbjct: 915 RSAEVFDVATWKWGPVEEEFMDAAACPRTFVDGGDGAMYMCCGGDVVALQGDTWRNVAKV 1094
Query: 298 PSEIRNVACMGA 309
P EIR G+
Sbjct: 1095PGEIRXRCVRGS 1130
Score = 95.9 bits (237), Expect(2) = e-160
Identities = 42/56 (75%), Positives = 50/56 (89%)
Frame = +1
Query: 304 VACMGAWERSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQSYCLLEI 359
VAC+G WE ++L+IGSSGFGEPH+GF+LDLKS W KLVSP++YTGHVQS CLLEI
Sbjct: 1114 VACVGVWEGAMLVIGSSGFGEPHVGFVLDLKSWAWTKLVSPEEYTGHVQSGCLLEI 1281
>TC227628 similar to UP|Q6H8E0 (Q6H8E0) Kelch repeat-containing F-box-like,
partial (19%)
Length = 1524
Score = 451 bits (1161), Expect = e-127
Identities = 223/361 (61%), Positives = 273/361 (74%), Gaps = 2/361 (0%)
Frame = +3
Query: 1 MELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTA 60
MELISGLPEDVARDCLIRVSY+QFP V + CK W +EI +PEF R+RR T +TQK++
Sbjct: 210 MELISGLPEDVARDCLIRVSYQQFPTVASVCKLWKSEIHAPEFHRQRRSTKHTQKVIAMV 389
Query: 61 QARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGY 120
QA + T VK NPVY LSVFEPETGNWS++ PPEF SG LPMFCQ+ VGY
Sbjct: 390 QAHVEPGTGSTKRVK---NPVYWLSVFEPETGNWSKIPPPPEFYSG--LPMFCQLVSVGY 554
Query: 121 ELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHD 180
+LVV+GG DP SW+ASNSVF+YNFLSA WRRG DMPGG R FF+C SD + TV+VAGGHD
Sbjct: 555 DLVVLGGLDPNSWEASNSVFVYNFLSAKWRRGTDMPGGRRMFFSCASDSEGTVFVAGGHD 734
Query: 181 EEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERS 240
EKNAL+SALAYDV++D WV LPDMA ERDECK VF G+ VGGY TE QGRF +S
Sbjct: 735 NEKNALRSALAYDVSSDRWVVLPDMAAERDECKGVF--SRGRFVAVGGYPTETQGRFVKS 908
Query: 241 AEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGD--AMYMCRGGDVVALQGNTWQTVAKVP 298
AE FD T W V ++FLDC TCPRT +DGGD +++C G D++AL+G+TWQ +A +P
Sbjct: 909 AEAFDPATSSWSEV-KDFLDCATCPRTFVDGGDDEGVFLCSGEDLIALRGDTWQKMASLP 1085
Query: 299 SEIRNVACMGAWERSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQSYCLLE 358
EIRNV+ +GA++ +L+LIGSSG+GE H+ + D+KS KL SP+ + GHVQ+ C+LE
Sbjct: 1086GEIRNVSYVGAFDGTLVLIGSSGYGEVHVAYAFDVKSCNGRKLESPEGFRGHVQTGCVLE 1265
Query: 359 I 359
I
Sbjct: 1266I 1268
>BQ612041
Length = 417
Score = 198 bits (504), Expect = 3e-51
Identities = 91/119 (76%), Positives = 104/119 (86%), Gaps = 1/119 (0%)
Frame = +3
Query: 231 TEMQGRFERSAEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGD-AMYMCRGGDVVALQGN 289
TEMQGRFERSAE FDV TW+WGPVEEEF+D CPRT +DGGD AMYMC GGDVVALQG+
Sbjct: 60 TEMQGRFERSAEVFDVATWKWGPVEEEFMDAAACPRTFVDGGDGAMYMCCGGDVVALQGD 239
Query: 290 TWQTVAKVPSEIRNVACMGAWERSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYT 348
TW+ VAKVP EIRNVAC+G WE ++L+IGSSGFGEPH+GF+LDLKS W KLVSP++YT
Sbjct: 240 TWRNVAKVPGEIRNVACVGVWEGAMLVIGSSGFGEPHVGFVLDLKSWAWTKLVSPEEYT 416
>TC209382
Length = 1333
Score = 125 bits (314), Expect(2) = 3e-51
Identities = 71/178 (39%), Positives = 103/178 (56%), Gaps = 2/178 (1%)
Frame = +3
Query: 2 ELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQ 61
+LI LP ++ +CL R+ + C W+ +QS F R+ TG+T+K+ Q
Sbjct: 162 DLIPKLPSELGLECLTRLPHSAHRVALRVCSQWHCLLQSDAFYSHRKKTGHTRKVTCLVQ 341
Query: 62 ARFDSDTCGGLLVK--ATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVG 119
AR D L K A+ VY +SVF+PE+ W + P++ SG LP+FCQ+A
Sbjct: 342 AREDQP----LQEKNNASVASVYGISVFDPESMTWDRVDPVPDYPSG--LPLFCQLASCD 503
Query: 120 YELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAG 177
+LV+MGGWDP S++ +VF+Y+F ++ WRRG DMP R+FFA + R VYVAG
Sbjct: 504 GKLVLMGGWDPASYEPLTAVFVYDFRTSEWRRGKDMP-EKRSFFAIGAGVGR-VYVAG 671
Score = 94.7 bits (234), Expect(2) = 3e-51
Identities = 70/184 (38%), Positives = 93/184 (50%), Gaps = 1/184 (0%)
Frame = +1
Query: 177 GGHDEEKNALKSALAYDVANDEWVPLPDMA-RERDECKAVFRGGAGKLRVVGGYCTEMQG 235
GGHDE KNAL +A AYD +DEW L M RE K V G + VV GY TE QG
Sbjct: 670 GGHDENKNALSTAWAYDPRSDEWAGLDPMGPREGTSAKGVVIG--DEFWVVSGYSTERQG 843
Query: 236 RFERSAEEFDVDTWQWGPVEEEFLDCGTCPRTCLDGGDAMYMCRGGDVVALQGNTWQTVA 295
F+ SAE D+ + W E F + G CPR+C+ G + G +V +G W
Sbjct: 844 MFDGSAEVLDIGSGGWRE-ENGFWEEGQCPRSCVGVG------KDGRLVNWRG--WDPGL 996
Query: 296 KVPSEIRNVACMGAWERSLLLIGSSGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQSYC 355
+V C A ++L GS G PH +L+D + K K+ P+ ++G VQS C
Sbjct: 997 RVG------VCGVAVGSRVVLSGSDYEGAPHGFYLVD--NRKLTKINVPEGFSGFVQSGC 1152
Query: 356 LLEI 359
+EI
Sbjct: 1153CVEI 1164
>TC227629 similar to UP|Q6H8E0 (Q6H8E0) Kelch repeat-containing F-box-like,
partial (17%)
Length = 632
Score = 189 bits (479), Expect = 2e-48
Identities = 92/160 (57%), Positives = 118/160 (73%), Gaps = 2/160 (1%)
Frame = +1
Query: 202 LPDMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERSAEEFDVDTWQWGPVEEEFLDC 261
LPDM ERDECK VF G+ VGGY TE QGRF +SAE FD T W V+E+FLDC
Sbjct: 4 LPDMEAERDECKGVFC--RGRFVAVGGYPTETQGRFVKSAEAFDPATRSWSEVKEDFLDC 177
Query: 262 GTCPRTCLDGGD--AMYMCRGGDVVALQGNTWQTVAKVPSEIRNVACMGAWERSLLLIGS 319
TCPRT +DGGD +++C GGD++AL+G+TWQ +A +P EIRNVA +GA++ +L+LIGS
Sbjct: 178 ATCPRTFVDGGDDEGVFLCSGGDLMALRGDTWQMMATLPGEIRNVAYVGAFDGTLVLIGS 357
Query: 320 SGFGEPHMGFLLDLKSGKWAKLVSPQDYTGHVQSYCLLEI 359
SG+GE H+ F D+KS W KL SP+ + GHVQ+ C+LEI
Sbjct: 358 SGYGEVHVSFAFDVKSCCWRKLESPEGFRGHVQTGCVLEI 477
>TC229125 weakly similar to GB|AAC13686.1|3047308|AF056929 sarcosin {Homo
sapiens;} , partial (4%)
Length = 1454
Score = 69.3 bits (168), Expect = 3e-12
Identities = 61/225 (27%), Positives = 95/225 (42%), Gaps = 8/225 (3%)
Frame = +2
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
LI LP+ VA CL RV + P + + W I+SPE + R+ G+T+ +L
Sbjct: 182 LIERLPDAVAIRCLARVPFYFHPVLELVSRSWQAAIRSPELFKARQEVGSTEDLLCV--C 355
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPP-------EFDSGSGLPMFCQI 115
FD P +++P W L + P + S +I
Sbjct: 356 AFD--------------PENLWQLYDPMRDLWITLPVLPSKIRHLSNLGAVSTAGKLFEI 493
Query: 116 AGVGYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYV 175
G G + V D + A++ V+ Y+ ++ W A M PR+ FAC + V V
Sbjct: 494 GG-GSDAVDPLTGDQDGCFATDEVWSYDPVAREWASRASML-VPRSMFACCVLNGKIV-V 664
Query: 176 AGGHDEEKNALKSALAYDVANDEWVPLPDMARERDE-CKAVFRGG 219
AGG + ++ + YD D W+P+PD+ R + C V GG
Sbjct: 665 AGGFTSCRKSISQSEMYDPDKDIWIPMPDLHRTHNSACSGVVIGG 799
>TC208758 similar to UP|Q8L736 (Q8L736) Predicted protein, partial (52%)
Length = 797
Score = 58.9 bits (141), Expect = 3e-09
Identities = 52/175 (29%), Positives = 71/175 (39%), Gaps = 3/175 (1%)
Frame = +2
Query: 87 FEPETGNWSELTMPPEFDSGSGLPMFC---QIAGVGYELVVMGGWDPESWKASNSVFIYN 143
F+P T W L P + C + VG EL+V G S ++ Y+
Sbjct: 137 FDPNTRRWMRLPRMPSNEC-----FICSDKESLAVGTELLVFG-----KEIMSPVIYRYS 286
Query: 144 FLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHDEEKNALKSALAYDVANDEWVPLP 203
L W G +M PR F S + + +AGG D N L SA Y+ W LP
Sbjct: 287 ILMNAWSSGMEM-NIPRCLFGSASLGEIAI-LAGGCDPRGNILSSAELYNSETGTWELLP 460
Query: 204 DMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERSAEEFDVDTWQWGPVEEEF 258
+M + R C VF GK V+GG + EEFD+ T +W + F
Sbjct: 461 NMNKARKMCSGVFID--GKFYVIGGIGVGNSKQL-TCGEEFDLQTRKWQKIPNMF 616
>TC217845 similar to UP|Q6K649 (Q6K649) Kelch repeat-containing F-box
protein-like, partial (71%)
Length = 1049
Score = 57.0 bits (136), Expect = 1e-08
Identities = 40/110 (36%), Positives = 57/110 (51%), Gaps = 2/110 (1%)
Frame = +3
Query: 121 ELVVMGGWDPESWKASNSVFIYNFLSA--TWRRGADMPGGPRTFFACTSDQDRTVYVAGG 178
+L+VM G+ AS S +Y + S +W R + M R FAC ++ D VY GG
Sbjct: 108 KLLVMAGYSSIDGTASVSAEVYQYDSCLNSWSRLSSM-NVARYDFAC-AEVDGLVYAVGG 281
Query: 179 HDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGG 228
+ ++L SA YD+ D+W P+ + R R C A G GKL V+GG
Sbjct: 282 YGATGDSLSSAEVYDLDTDKWTPIESLRRPRWGCFAC--GFEGKLYVMGG 425
Score = 33.9 bits (76), Expect = 0.12
Identities = 33/105 (31%), Positives = 44/105 (41%), Gaps = 4/105 (3%)
Frame = +3
Query: 155 MPGGPRTFFACTSDQDRTVYVAGGHDEEKNALKSA--LAYDVANDEWVPLPDM--ARERD 210
MPG + F + + +AG + A SA YD + W L M AR
Sbjct: 60 MPGPAKAGFGVVVLNGKLLVMAGYSSIDGTASVSAEVYQYDSCLNSWSRLSSMNVARYDF 239
Query: 211 ECKAVFRGGAGKLRVVGGYCTEMQGRFERSAEEFDVDTWQWGPVE 255
C V G + VGGY G SAE +D+DT +W P+E
Sbjct: 240 ACAEV----DGLVYAVGGYGAT--GDSLSSAEVYDLDTDKWTPIE 356
>TC211400 similar to UP|Q9FKJ0 (Q9FKJ0) Emb|CAB55405.1, partial (52%)
Length = 767
Score = 55.1 bits (131), Expect = 5e-08
Identities = 59/231 (25%), Positives = 85/231 (36%), Gaps = 5/231 (2%)
Frame = +2
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
LI GL +DVA +CL + ++ K +N I S R+ G + ++
Sbjct: 143 LIPGLIDDVALNCLAWFCGSDYAVLSCINKRFNKLINSGYLYGLRKQLGAVEHLVYMV-- 316
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPP-----EFDSGSGLPMFCQIAG 117
D G + F+P+ W L P L + C++
Sbjct: 317 ---CDPRGWV-------------AFDPKINRWISLPKIPCDECFNHADKESLAVGCELLV 448
Query: 118 VGYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAG 177
G EL+ W Y+ + W + +M PR F +S VAG
Sbjct: 449 FGRELMEFAIWK------------YSMICRGWVKCQEM-NQPRCLFG-SSSLGSIAIVAG 586
Query: 178 GHDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGG 228
G D+ N LKSA YD + W LP+M R C F GK V+GG
Sbjct: 587 GSDKYGNVLKSAELYDSSTGMWELLPNMHAPRRLCSGFFMD--GKFYVIGG 733
>TC227618 similar to UP|Q9FKJ0 (Q9FKJ0) Emb|CAB55405.1, partial (73%)
Length = 1190
Score = 53.1 bits (126), Expect = 2e-07
Identities = 53/175 (30%), Positives = 73/175 (41%), Gaps = 2/175 (1%)
Frame = +2
Query: 86 VFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYELVVMGG--WDPESWKASNSVFIYN 143
VF+P+ W L P D +AG G E++V G D WK Y+
Sbjct: 44 VFDPKKNRWITLPKIP-CDECFNHADKESLAG-GKEMLVFGREMMDFAIWK-------YS 196
Query: 144 FLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHDEEKNALKSALAYDVANDEWVPLP 203
+S W + +M PR F S + VAGG D+ N L+SA YD + W LP
Sbjct: 197 LISRGWVKCKEM-NHPRCLFGSGSLGSIAI-VAGGSDKYGNVLESAELYDSNSGTWKLLP 370
Query: 204 DMARERDECKAVFRGGAGKLRVVGGYCTEMQGRFERSAEEFDVDTWQWGPVEEEF 258
+M R C F GK V+GG + EE+D+ T W +E +
Sbjct: 371 NMHTPRRLCSGFFMD--GKFYVIGGMSSPTVSL--TCGEEYDLKTRNWRKIERMY 523
>TC229968 similar to UP|O64797 (O64797) T1F15.5 protein, partial (13%)
Length = 902
Score = 51.6 bits (122), Expect = 5e-07
Identities = 24/55 (43%), Positives = 33/55 (59%)
Frame = +3
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKIL 57
++ GLP+DVA CL V FPA+ CKGW + IQS EF R++ G ++ L
Sbjct: 708 ILPGLPDDVAEYCLALVPRSNFPAMGVVCKGWRSFIQSKEFTTVRKLAGMLEEWL 872
>TC208333 weakly similar to GB|AAC13686.1|3047308|AF056929 sarcosin {Homo
sapiens;} , partial (6%)
Length = 909
Score = 47.4 bits (111), Expect = 1e-05
Identities = 30/86 (34%), Positives = 43/86 (49%), Gaps = 1/86 (1%)
Frame = +3
Query: 135 ASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGGHDEEKNALKSALAYDV 194
A+N V+ Y+ + W A M PR+ FAC + V VAGG + ++ A YD
Sbjct: 9 ATNEVWSYDPVVRQWSPRAAMLV-PRSMFACCVMNGKIV-VAGGFTSCRKSISQAEMYDP 182
Query: 195 ANDEWVPLPDMARERDE-CKAVFRGG 219
D W+P+PD+ R + C V GG
Sbjct: 183 EKDVWIPMPDLHRTHNSACSGVVIGG 260
>TC221969
Length = 581
Score = 45.1 bits (105), Expect = 5e-05
Identities = 41/142 (28%), Positives = 62/142 (42%), Gaps = 1/142 (0%)
Frame = +1
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
L+SGLP+D+A CLIRV + + CK W+ + F R+ G ++ L +A
Sbjct: 193 LLSGLPDDLAIACLIRVPRIEHRKLHLVCKRWHRLLSEDFFYSLRKSLGMAEEWLYVIKA 372
Query: 63 RFDSDTCGGLLVKATTNPVYRL-SVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYE 121
D G + V A +P+Y+L P G++ P GS + C
Sbjct: 373 ----DRAGRISVHA-FDPIYQLWQPLPPVPGDF-----PEAMWVGSAVLSGC-------H 501
Query: 122 LVVMGGWDPESWKASNSVFIYN 143
L + GG D E ++ V YN
Sbjct: 502 LYLFGGVDLEGSRSIRRVIFYN 567
>AW186414
Length = 445
Score = 37.4 bits (85), Expect = 0.011
Identities = 17/55 (30%), Positives = 29/55 (51%)
Frame = +1
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKIL 57
LI GLP+ VA CL V + P + + W ++ PE + R+ G+++ +L
Sbjct: 208 LIEGLPDAVAIRCLAWVPFYLHPKLELVSRAWRAVVRGPELFKARQELGSSEDLL 372
>AW596139 weakly similar to GP|18086559|gb| At2g24540/F25P17.16 {Arabidopsis
thaliana}, partial (22%)
Length = 411
Score = 34.7 bits (78), Expect = 0.069
Identities = 31/126 (24%), Positives = 46/126 (35%)
Frame = +1
Query: 2 ELISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQ 61
ELI GLP ++A CL+ V Y + WN I P F ++ ++
Sbjct: 28 ELIPGLPYEIAELCLLHVPYPYQALSRSVSSTWNRAITHPSFIFSKKTLSRPHLFVLA-- 201
Query: 62 ARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYE 121
F S T G + +A +P +G W L P ++ C +
Sbjct: 202 --FHSQT-GKIQWQA----------LDPSSGRWFVLPQMPLQENSCPTEFACAALPHQGK 342
Query: 122 LVVMGG 127
L VM G
Sbjct: 343 LFVMAG 360
>TC219334 weakly similar to UP|Q9FUZ9 (Q9FUZ9) SKP1 interacting partner 6
(Fragment), partial (29%)
Length = 1451
Score = 33.5 bits (75), Expect = 0.15
Identities = 51/211 (24%), Positives = 79/211 (37%), Gaps = 4/211 (1%)
Frame = +3
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPEFRRRRRVTGNTQKILVTAQA 62
LI LP+DVA +CL + P ++ K +T + SP R + TQ +L
Sbjct: 60 LIPSLPDDVALNCLGHIPQSXHPTLSLISKPIHTLLSSPILFTTRTLLQCTQPLLY---- 227
Query: 63 RFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELTMPPEFDSGSGLPMFCQIAGVGYEL 122
L + + + + + N LT P S + + A +G +
Sbjct: 228 ---------LTLHSHHSSLLQFFTLHHTNPNNPLLTPLPPIPSPA---VGSAYAVLGPTI 371
Query: 123 VVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFACTSDQDRTVYVAGG--HD 180
V+GG + S +V++ + W RG M G FA +YV GG D
Sbjct: 372 YVLGGSIHD--VPSPNVWLLDCRFNRWLRGPSMRVGRE--FAAAGVLHGKIYVLGGCVAD 539
Query: 181 EEKNALKSALAYDVANDEW--VPLPDMARER 209
+ A D A +W V P RE+
Sbjct: 540 TWSRSANWAEVLDPATGQWERVASPTEVREK 632
Score = 28.1 bits (61), Expect = 6.4
Identities = 25/82 (30%), Positives = 33/82 (39%), Gaps = 2/82 (2%)
Frame = +3
Query: 172 TVYVAGG--HDEEKNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGGY 229
T+YV GG HD + D + W+ P M R+ A G K+ V+GG
Sbjct: 366 TIYVLGGSIHDVPS---PNVWLLDCRFNRWLRGPSMRVGREFAAAGVLHG--KIYVLGGC 530
Query: 230 CTEMQGRFERSAEEFDVDTWQW 251
+ R AE D T QW
Sbjct: 531 VADTWSRSANWAEVLDPATGQW 596
>TC210605 weakly similar to UP|Q9FV01 (Q9FV01) SKP1 interacting partner 4,
partial (12%)
Length = 661
Score = 32.3 bits (72), Expect = 0.34
Identities = 36/168 (21%), Positives = 68/168 (40%), Gaps = 5/168 (2%)
Frame = +1
Query: 3 LISGLPEDVARDCLIRVSYEQFPAVTAACKGWNTEIQSPE---FRRRRRVTGNTQKILVT 59
+I GLP+D++ CL R+ + + K W I S E +RR+ ++
Sbjct: 199 IICGLPDDISLMCLARIPRKYHSVMKCVSKRWRNLICSEEWFCYRRKHKL---------- 348
Query: 60 AQARFDSDTCGGLLVKATTNPVYRLSVFEPETGNWSELT--MPPEFDSGSGLPMFCQIAG 117
+T L + +N ++ + + + +L +PP G+
Sbjct: 349 ------DETWIYALCRDKSNEIFCYVLDPTDPIRYWKLVGGLPPHISKREGM----GFEV 498
Query: 118 VGYELVVMGGWDPESWKASNSVFIYNFLSATWRRGADMPGGPRTFFAC 165
+G +L ++GG E ++N V+ Y+ S W + + R FAC
Sbjct: 499 LGNKLFLLGGC-REFLGSTNEVYSYDASSNCWAQATSL-STARYNFAC 636
>BE804114
Length = 159
Score = 31.6 bits (70), Expect = 0.58
Identities = 18/46 (39%), Positives = 25/46 (54%)
Frame = +2
Query: 183 KNALKSALAYDVANDEWVPLPDMARERDECKAVFRGGAGKLRVVGG 228
+++L SA YD D+W + + R R C A G GKL V+GG
Sbjct: 14 RDSLSSAEVYDPDTDKWALIESLRRPRWGCFAC--GFEGKLYVMGG 145
>TC213795
Length = 631
Score = 30.0 bits (66), Expect = 1.7
Identities = 21/71 (29%), Positives = 33/71 (45%), Gaps = 4/71 (5%)
Frame = +2
Query: 194 VANDEWVPLPDMARERDECKAVFRGGAG----KLRVVGGYCTEMQGRFERSAEEFDVDTW 249
V D+WV P ++ +R KA + GA KL + GG GR+ D+ +W
Sbjct: 20 VVYDQWVA-PPVSGQRP--KARYEHGAAVVQDKLYIYGG---NHNGRYLNDLHVLDLRSW 181
Query: 250 QWGPVEEEFLD 260
W +E E ++
Sbjct: 182 TWSKIEAEVVE 214
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,468,778
Number of Sequences: 63676
Number of extensions: 284026
Number of successful extensions: 1189
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 1151
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1163
length of query: 359
length of database: 12,639,632
effective HSP length: 98
effective length of query: 261
effective length of database: 6,399,384
effective search space: 1670239224
effective search space used: 1670239224
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0253.11


