

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0253.10
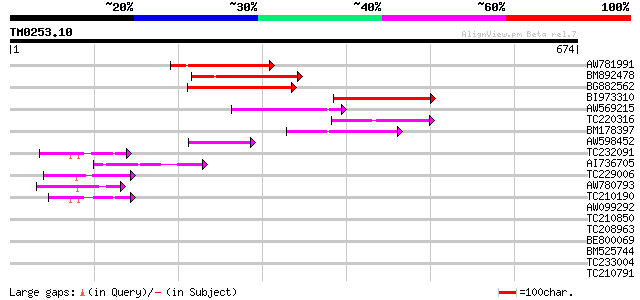
(674 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW781991 160 1e-39
BM892478 154 2e-37
BG882562 135 4e-32
BI973310 106 3e-23
AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis... 99 5e-21
TC220316 94 3e-19
BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thalia... 84 2e-16
AW598452 weakly similar to GP|15982769|gb| At2g27110/T20P8.16 {A... 76 4e-14
TC232091 59 7e-09
AI736705 similar to GP|11994736|db far-red impaired response pro... 52 9e-07
TC229006 52 1e-06
AW780793 46 6e-05
TC210190 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, part... 42 7e-04
AW099292 41 0.002
TC210850 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, part... 39 0.006
TC208963 37 0.022
BE800069 34 0.24
BM525744 34 0.24
TC233004 weakly similar to PIR|G96565|G96565 F6D8.26 [imported] ... 33 0.31
TC210791 33 0.41
>AW781991
Length = 419
Score = 160 bits (406), Expect = 1e-39
Identities = 70/123 (56%), Positives = 93/123 (74%)
Frame = +3
Query: 192 FTNRDCWNYIRNVRRKNLDVGDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDA 251
FT DC NY+RN R ++L+ GD Q V DY ++ EN NFFYA+Q DED + N+FW D
Sbjct: 54 FTEVDCRNYMRNNRLRSLE-GDIQLVLDYLRQMHAENPNFFYAVQGDEDQSITNVFWADP 230
Query: 252 RSRLSYQLFGDVITFDTTYKTNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLF 311
++R++Y FGD +TFDTTY++N+Y +PFA F G+N+H Q +L+GCA L +ESEASFVWLF
Sbjct: 231 KARMNYTFFGDTVTFDTTYRSNRYRLPFAFFTGVNHHGQPVLFGCAFLINESEASFVWLF 410
Query: 312 KTW 314
KTW
Sbjct: 411 KTW 419
>BM892478
Length = 429
Score = 154 bits (388), Expect = 2e-37
Identities = 68/132 (51%), Positives = 97/132 (72%)
Frame = +3
Query: 217 VFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNKYS 276
+ +Y + +Q E++ FFYA++ D + M N+FW D RSR S FGDV+ DT+Y+ Y
Sbjct: 30 LLEYFQSRQAEDTGFFYAMEVDNGNCM-NIFWADGRSRYSCSHFGDVLVLDTSYRKTVYL 206
Query: 277 MPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQDLAMKA 336
+PFA FVG+N+H Q +L GCAL+ DESE SF WLF+TWL+AM G+ P+++I DQD+A++
Sbjct: 207 VPFATFVGVNHHKQPVLLGCALIADESEESFTWLFQTWLRAMSGRLPLTVIADQDIAIQR 386
Query: 337 ALAKVFPESRHR 348
A+AKVFP + HR
Sbjct: 387 AIAKVFPVTHHR 422
>BG882562
Length = 416
Score = 135 bits (341), Expect = 4e-32
Identities = 58/130 (44%), Positives = 91/130 (69%)
Frame = +3
Query: 212 GDAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYK 271
GD + + +Y R Q+ N NFFY + ++D ++ N+FW+D+RSR +Y FGDV+ FD+T
Sbjct: 27 GDPELISNYFCRIQLMNPNFFYVMDLNDDGQLRNVFWIDSRSRAAYSYFGDVVAFDSTCL 206
Query: 272 TNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKPISIITDQD 331
+N Y +P FVG+N+H +S+L GC LL DE+ +++WLF+ WL M G+ P +IIT+
Sbjct: 207 SNNYEIPLVAFVGVNHHGKSVLLGCGLLADETFETYIWLFRAWLTCMTGRPPQTIITNXC 386
Query: 332 LAMKAALAKV 341
AM++A+A+V
Sbjct: 387 KAMQSAIAEV 416
>BI973310
Length = 457
Score = 106 bits (265), Expect = 3e-23
Identities = 49/122 (40%), Positives = 78/122 (63%)
Frame = +3
Query: 385 SIQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFF 444
+I FE W ++ + + +NEWLQ +Y R+ W+P+Y R TFF ++ + +E + +FF
Sbjct: 3 TIDEFESYWHSLLERFYVMDNEWLQSIYNSRQHWVPVYLRETFFGEISLNEGNEYLISFF 182
Query: 445 DSFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEI 504
D +V +STTLQ V ++EKAV S E E K +Y++ + S +L T S +E+ A LYT +I
Sbjct: 183 DGYVFSSTTLQVLVRQYEKAVSSWHEKELKADYDTTNSSPVLKTPSPMEKQAASLYTRKI 362
Query: 505 FL 506
F+
Sbjct: 363 FM 368
>AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana},
partial (19%)
Length = 425
Score = 99.4 bits (246), Expect = 5e-21
Identities = 46/138 (33%), Positives = 81/138 (58%), Gaps = 1/138 (0%)
Frame = +1
Query: 264 ITFDTTYKTNKYSMPFAPFVGLNNHSQSILYGCALLQDESEASFVWLFKTWLQAMGGKKP 323
+ FDTTY+ Y M ++G++N+ + + CALL+DE+ SF W K +L M GK P
Sbjct: 10 VVFDTTYRVEAYDMLLGIWLGVDNNGMTCFFSCALLRDENIQSFSWALKAFLGFMKGKAP 189
Query: 324 ISIITDQDLAMKAALAKVFPESRHRLCLWHIIKKFPEKLAHIYHKQ-SIFKRDMKRCIRG 382
+I+TD ++ +K A+A P+++H C+WHI+ KF + + + Q +K + R +
Sbjct: 190 QTILTDHNMWLKEAIAVELPQTKHAFCIWHILSKFSDWFSLLLGSQYDEWKAEFHR-LYN 366
Query: 383 SHSIQSFEEEWMRIMVEY 400
++ FEE W +++ +Y
Sbjct: 367 LEQVEDFEEGWRQMVDQY 420
>TC220316
Length = 988
Score = 93.6 bits (231), Expect = 3e-19
Identities = 44/123 (35%), Positives = 74/123 (59%)
Frame = +1
Query: 383 SHSIQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINA 442
S ++ W +Y LK++ WL+ +Y+ R SW+P+Y + TFFAG+ +ES+++
Sbjct: 16 SQTVDEXXATWNXXXNKYGLKDDAWLKEMYQKRASWVPLYLKGTFFAGI---PMNESLDS 186
Query: 443 FFDSFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTP 502
FF + ++A T L EF+ ++E+ +E R E ERKE++ + + IL T +EE LYT
Sbjct: 187 FFGALLNAQTPLMEFIPRYERGLERRREEERKEDFNTSNFQPILQTKEPVEEQCRRLYTL 366
Query: 503 EIF 505
+F
Sbjct: 367 TVF 375
>BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana}, partial
(20%)
Length = 422
Score = 84.3 bits (207), Expect = 2e-16
Identities = 47/138 (34%), Positives = 72/138 (52%), Gaps = 1/138 (0%)
Frame = +1
Query: 330 QDLAMKAALAKVFPESRHRLCLWHIIKKFPEKL-AHIYHKQSIFKRDMKRCIRGSHSIQS 388
Q++ +K AL+ P ++H C+W I+ KFP A + + + +K + R + S++
Sbjct: 1 QNICLKEALSTEMPTTKHAFCIWMIVAKFPSWFNAVLGERYNDWKAEFYR-LYNLESVED 177
Query: 389 FEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNTTQRSESINAFFDSFV 448
FE W + + L N + LY R W + RS F AGM TT +S+SINAF F+
Sbjct: 178 FELGWREMACSFGLHSNRHMVNLYSSRSLWALPFLRSHFLAGMTTTGQSKSINAFIQRFL 357
Query: 449 DASTTLQEFVLKFEKAVE 466
A T L FV + AV+
Sbjct: 358 SAQTRLAHFVEQVAVAVD 411
>AW598452 weakly similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidopsis
thaliana}, partial (7%)
Length = 256
Score = 76.3 bits (186), Expect = 4e-14
Identities = 35/80 (43%), Positives = 48/80 (59%)
Frame = +2
Query: 213 DAQAVFDYCKRKQVENSNFFYAIQCDEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKT 272
DA + +YCKR Q EN FYAIQ DED R+ N+ RSR SY+ GD + D Y
Sbjct: 17 DAHNLLEYCKRMQAENHGCFYAIQLDEDDRVSNVL*AGTRSRTSYKHMGDAESLDAAYGK 196
Query: 273 NKYSMPFAPFVGLNNHSQSI 292
++Y++ +AP+ G N H + I
Sbjct: 197 HRYAVSYAPYTGRNCHGRMI 256
>TC232091
Length = 530
Score = 58.9 bits (141), Expect = 7e-09
Identities = 34/117 (29%), Positives = 59/117 (50%), Gaps = 7/117 (5%)
Frame = +1
Query: 36 GELNSKPEIGMEFESIEKVREFYNSFDKKNGFGVRV----RSSKPKRAV---LVCCNEGQ 88
G +P +GMEF+S E + FY + ++ GF VR+ RS K+ + C +G
Sbjct: 118 GSSIEEPNVGMEFQSEEAAKNFYEEYARREGFVVRLDRCHRSEVDKQIISRRFSCNKQGF 297
Query: 89 HKVKISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVM 145
H +++ T + ++IR GCEA + V + T KW++ F +H+H++
Sbjct: 298 H-------VRVRNKTKPVHKPRASIREGCEAMMYV-KVNTCGKWVVTKFVKEHSHLL 444
>AI736705 similar to GP|11994736|db far-red impaired response protein;
Mutator-like transposase-like protein; phytochrome A
signaling, partial (14%)
Length = 383
Score = 52.0 bits (123), Expect = 9e-07
Identities = 40/137 (29%), Positives = 65/137 (47%), Gaps = 1/137 (0%)
Frame = +3
Query: 100 KDSTNQT-KRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPKSVSYMRCHK 158
+DS N T +R CS ++ C+AS+ V R + KW+I SF +HNH ++ ++VS + +
Sbjct: 39 QDSENSTGRRSCS--KTDCKASMHVKR-RSDGKWVIHSFVKEHNHELLPAQAVSE-QTRR 206
Query: 159 KMSVAAKSLVEKFEEEGIPTGKVAAIFNDGDSTFTNRDCWNYIRNVRRKNLDVGDAQAVF 218
+ A+ E G+ K N R L+ G+A+ +
Sbjct: 207 MYAAMARQFAEYKTVVGLKNEK------------------NPFDKGRNLGLESGEAKLML 332
Query: 219 DYCKRKQVENSNFFYAI 235
D+ + Q NSNFFYA+
Sbjct: 333 DFFIQMQNMNSNFFYAV 383
>TC229006
Length = 991
Score = 51.6 bits (122), Expect = 1e-06
Identities = 37/116 (31%), Positives = 62/116 (52%), Gaps = 7/116 (6%)
Frame = +2
Query: 41 KPEIGMEFESIEKVREFYNSFDKKNGFGVRVRS-SKPKR-----AVLVCCNEGQHKVKIS 94
+P GMEFES + + FY+ + ++ GF +RV S +P+R A + CN+ + V
Sbjct: 98 EPYEGMEFESEDAAKIFYDEYARRLGFVMRVMSCRRPERDGRILARRLGCNKEGYCV--- 268
Query: 95 RTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNH-VMVSPK 149
I+ ++ ++ R GC+A +I + KW+I F DHNH ++VSP+
Sbjct: 269 ---SIRGKFASVRKPRASTREGCKA-MIHIKYNKSGKWVITKFVKDHNHPLVVSPR 424
>AW780793
Length = 419
Score = 45.8 bits (107), Expect = 6e-05
Identities = 33/115 (28%), Positives = 57/115 (48%), Gaps = 9/115 (7%)
Frame = +3
Query: 32 SDIGGELNS-KPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKRA-------VLVC 83
+DI ++ + +P IGMEF S E+ REFY ++ ++ GF VR+ ++ R VC
Sbjct: 99 ADISTDIEAVEPFIGMEFNSREEAREFYIAYGRRIGFTVRIHHNRRSRVNNQVIGQDFVC 278
Query: 84 CNEG-QHKVKISRTEEIKDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSF 137
EG + K + R + + T R GC+A + ++ + KW++ F
Sbjct: 279 SKEGFRAKKYLHRKDRVLPPPPAT-------REGCQAMIRLAL-RDRGKWVVTKF 419
>TC210190 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, partial (65%)
Length = 866
Score = 42.4 bits (98), Expect = 7e-04
Identities = 33/110 (30%), Positives = 49/110 (44%), Gaps = 7/110 (6%)
Frame = +1
Query: 47 EFESIEKVREFYNSFDKKNGFGVRV----RSSKPKRAV---LVCCNEGQHKVKISRTEEI 99
EF S FYN++ + GF VRV RS + A+ LVC EG +
Sbjct: 4 EFGSEAAAHAFYNAYATEVGFIVRVSKLSRSRRDGTAIGRTLVCNKEGFR---------M 156
Query: 100 KDSTNQTKRKCSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPK 149
D + R+ + R GC A ++V R + KW++ F +H H + K
Sbjct: 157 ADKREKIVRQRAETRVGCRAMIMV-RKLSSGKWVVAKFVKEHTHPLTPGK 303
>AW099292
Length = 420
Score = 41.2 bits (95), Expect = 0.002
Identities = 22/52 (42%), Positives = 28/52 (53%)
Frame = +2
Query: 28 ILDTSDIGGELNSKPEIGMEFESIEKVREFYNSFDKKNGFGVRVRSSKPKRA 79
I+D+S + G S+P MEFES E FY + K GFG SS+ RA
Sbjct: 218 IMDSSAVVGSTISEPHNDMEFESHEAAYAFYKEYAKSAGFGTAKLSSRRSRA 373
>TC210850 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, partial (63%)
Length = 1326
Score = 39.3 bits (90), Expect = 0.006
Identities = 31/100 (31%), Positives = 45/100 (45%), Gaps = 7/100 (7%)
Frame = +1
Query: 57 FYNSFDKKNGFGVRV----RSSKPKRAV---LVCCNEGQHKVKISRTEEIKDSTNQTKRK 109
FYN++ GF VRV RS + A+ LVC EG + D + R+
Sbjct: 16 FYNAYATDVGFIVRVSKLSRSRRDGTAIGRTLVCNKEGFR---------MADKREKIVRQ 168
Query: 110 CSTIRSGCEASLIVSRGTTKSKWMIKSFNNDHNHVMVSPK 149
+ R GC A ++V R + KW+I F +H H + K
Sbjct: 169 RAETRVGCRAMIMV-RKLSSGKWVIAKFVKEHTHPLTPGK 285
>TC208963
Length = 1163
Score = 37.4 bits (85), Expect = 0.022
Identities = 35/156 (22%), Positives = 64/156 (40%), Gaps = 9/156 (5%)
Frame = +1
Query: 323 PISIITDQDLAMKAALAKVFPESRHRLCLWHI---IKKFPEKLAHIYHKQSIFKRDMKRC 379
P I+T D+A+ +A+ VFP S + LC +HI +K + + H+ KQ +
Sbjct: 22 PQVIVTVGDIALMSAVQVVFPSSSNLLCRFHINQNVKAKCKSIVHLKEKQELMMDAWDVV 201
Query: 380 IRGSHS------IQSFEEEWMRIMVEYNLKENEWLQGLYKIRESWIPIYNRSTFFAGMNT 433
+ + + FE + + + +N W L +E ++ + G
Sbjct: 202 VNSPNEGEYMQRLAFFENVCLDFPILGDYVKNTW---LIPHKEKFVTAWTNQVMHLGNTA 372
Query: 434 TQRSESINAFFDSFVDASTTLQEFVLKFEKAVESRL 469
T R +F+ F+ +T L KF A +R+
Sbjct: 373 TNRFAQ-TLYFE*FIRKNT*LIPHKEKFVTAWTNRV 477
>BE800069
Length = 233
Score = 33.9 bits (76), Expect = 0.24
Identities = 16/42 (38%), Positives = 27/42 (64%)
Frame = +1
Query: 465 VESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEIFL 506
+ESR E E + +Y++ + +L T S +E+ A LYT +IF+
Sbjct: 1 LESRNEKEVRADYDTMNTLPVLRTPSPMEKQASELYTRKIFM 126
>BM525744
Length = 411
Score = 33.9 bits (76), Expect = 0.24
Identities = 18/67 (26%), Positives = 30/67 (43%)
Frame = +3
Query: 238 DEDSRMVNLFWVDARSRLSYQLFGDVITFDTTYKTNKYSMPFAPFVGLNNHSQSILYGCA 297
D+ + ++FW + F V+ D TYK N+Y +P VG+ S + + A
Sbjct: 210 DDSDAIRDIFWAHPDAIKLLGSFHTVLFLDNTYKVNRYQLPLLEIVGVT--STELTFSVA 383
Query: 298 LLQDESE 304
ES+
Sbjct: 384 FAYMESD 404
>TC233004 weakly similar to PIR|G96565|G96565 F6D8.26 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(8%)
Length = 551
Score = 33.5 bits (75), Expect = 0.31
Identities = 19/61 (31%), Positives = 30/61 (49%)
Frame = +1
Query: 445 DSFVDASTTLQEFVLKFEKAVESRLEAERKENYESRHKSRILSTGSKLEEHAGHLYTPEI 504
D +V T+ +EFV K++ + + E + E+R+ S L T E +YT EI
Sbjct: 1 DGYVHKHTSFKEFVDKYDLVLHRKHLKEAMADLETRNVSFELKTRCNFEVQLAKVYTKEI 180
Query: 505 F 505
F
Sbjct: 181 F 183
>TC210791
Length = 456
Score = 33.1 bits (74), Expect = 0.41
Identities = 17/39 (43%), Positives = 23/39 (58%)
Frame = +1
Query: 323 PISIITDQDLAMKAALAKVFPESRHRLCLWHIIKKFPEK 361
P IIT +D+A+ A+ VFP S + LC +HI K K
Sbjct: 31 PPVIITVRDIALMDAVQVVFPSSSNLLCRFHISKNVKAK 147
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.333 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,786,693
Number of Sequences: 63676
Number of extensions: 402489
Number of successful extensions: 3176
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 3146
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3169
length of query: 674
length of database: 12,639,632
effective HSP length: 104
effective length of query: 570
effective length of database: 6,017,328
effective search space: 3429876960
effective search space used: 3429876960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0253.10


