

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252d.2
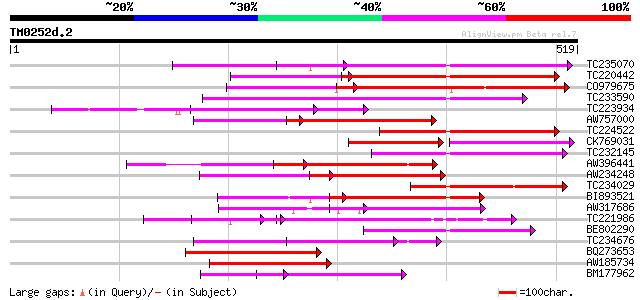
(519 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC235070 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 181 9e-46
TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 175 4e-44
CO979675 170 1e-42
TC233590 similar to UP|Q6K9T2 (Q6K9T2) Pentatricopeptide repeat-... 164 7e-41
TC223934 163 2e-40
AW757000 similar to PIR|C86440|C86 PPR-repeat protein [imported]... 139 2e-33
TC224522 weakly similar to UP|Q9LUL5 (Q9LUL5) Emb|CAB66100.1, pa... 129 4e-30
CK769031 80 6e-30
TC232145 128 7e-30
AW396441 127 9e-30
AW234248 122 3e-28
TC234029 similar to UP|Q9LN01 (Q9LN01) T6D22.15, partial (32%) 119 4e-27
BI893521 117 9e-27
AW317686 112 3e-25
TC221986 weakly similar to GB|BAC16401.1|23237826|AP003749 selen... 109 3e-24
BE802290 108 4e-24
TC234676 105 5e-23
BQ273653 103 2e-22
AW185734 101 9e-22
BM177962 101 9e-22
>TC235070 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (22%)
Length = 875
Score = 181 bits (458), Expect = 9e-46
Identities = 100/271 (36%), Positives = 148/271 (53%)
Frame = +2
Query: 245 ACAHLGALTQGLWVHSYAKRCKVDSNPILATALVDMYSKCGCVESALTVFEGIADKDAGA 304
AC+ L + QG +H++ + N + TALVD YSKCG + A F I + A
Sbjct: 2 ACSCLCSFRQGQLLHAHLIKTPFQVNVYVGTALVDFYSKCGHLAEAQRSFISIFSPNVAA 181
Query: 305 WNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGMVHQGLRLFEEMS 364
W A+I+G A G +++ LF M G P TFV VL+AC H G+V +GLR+F M
Sbjct: 182 WTALINGYAYHGLGSEAILLFRSMLHQGIVPNAATFVGVLSACNHAGLVCEGLRIFHSMQ 361
Query: 365 CTYGVEPRGEHYACVVDLLSRAGMVKEAEKFMEKMGGLVEADLNVWGAILNACRIYKNVD 424
YGV P EHY CVVDLL R+G +KEAE+F+ KM +EAD +WGA+LNA +K+++
Sbjct: 362 RCYGVTPTIEHYTCVVDLLGRSGHLKEAEEFIIKMP--IEADGIIWGALLNASWFWKDME 535
Query: 425 VGNRVWKKMIDKGVADCGTHILTYNMYREAGWDAEAKRVRSMVSEAGMKKKPGCSIIEVG 484
VG R +K+ ++ NMY G + ++R + ++K PGCS IE+
Sbjct: 536 VGERAAEKLFSLDPNPIFAFVVLSNMYAILGRWGQKTKLRKRLQSLELRKDPGCSWIELN 715
Query: 485 DEVEEFLAGDHSHPQAQELCKLLDSILMMAN 515
+++ F D +H + + ++ I N
Sbjct: 716 NKIHLFSVEDKTHLYSDVIYATVEHITATIN 808
Score = 65.9 bits (159), Expect = 4e-11
Identities = 44/165 (26%), Positives = 82/165 (49%), Gaps = 4/165 (2%)
Frame = +2
Query: 150 CASRQLQTARLLFDQTT-EKDVVLWTAMIDGYGKVGDVESARELFEEMPERNAVSWSAMM 208
C+ RQ Q +T + +V + TA++D Y K G + A+ F + N +W+A++
Sbjct: 17 CSFRQGQLLHAHLIKTPFQVNVYVGTALVDFYSKCGHLAEAQRSFISIFSPNVAAWTALI 196
Query: 209 AAYSRVSDFKQVLALFEEMQNRGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAKRCKVD 268
Y+ + + LF M ++G+ PN + V VL+AC H G + +GL + +RC
Sbjct: 197 NGYAYHGLGSEAILLFRSMLHQGIVPNAATFVGVLSACNHAGLVCEGLRIFHSMQRC-YG 373
Query: 269 SNPILA--TALVDMYSKCGCVESALT-VFEGIADKDAGAWNAMIS 310
P + T +VD+ + G ++ A + + + D W A+++
Sbjct: 374 VTPTIEHYTCVVDLLGRSGHLKEAEEFIIKMPIEADGIIWGALLN 508
>TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (25%)
Length = 706
Score = 175 bits (444), Expect = 4e-44
Identities = 85/200 (42%), Positives = 124/200 (61%)
Frame = +1
Query: 304 AWNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGMVHQGLRLFEEM 363
AW A+I G+A+ G R++L+ F QM G P TF A+LTAC+H G+ +G LFE M
Sbjct: 16 AWTAIIGGLAIHGKGREALDWFTQMQKAGINPNSITFTAILTACSHAGLTEEGKSLFESM 195
Query: 364 SCTYGVEPRGEHYACVVDLLSRAGMVKEAEKFMEKMGGLVEADLNVWGAILNACRIYKNV 423
S Y ++P EHY C+VDL+ RAG++KEA +F+E M V+ + +WGA+LNAC+++K+
Sbjct: 196 SSVYNIKPSMEHYGCMVDLMGRAGLLKEAREFIESMP--VKPNAAIWGALLNACQLHKHF 369
Query: 424 DVGNRVWKKMIDKGVADCGTHILTYNMYREAGWDAEAKRVRSMVSEAGMKKKPGCSIIEV 483
++G + K +I+ G +I ++Y AG + RVRS + G+ PGCS I +
Sbjct: 370 ELGKEIGKILIELDPDHSGRYIHLASIYAAAGEWNQVVRVRSQIKHRGLLNHPGCSSITL 549
Query: 484 GDEVEEFLAGDHSHPQAQEL 503
V EF AGD SHP QE+
Sbjct: 550 NGVVHEFFAGDGSHPHIQEI 609
Score = 45.8 bits (107), Expect = 5e-05
Identities = 27/114 (23%), Positives = 53/114 (45%), Gaps = 2/114 (1%)
Frame = +1
Query: 203 SWSAMMAAYSRVSDFKQVLALFEEMQNRGMKPNDSVLVTVLTACAHLGALTQGLWV-HSY 261
+W+A++ + ++ L F +MQ G+ PN +LTAC+H G +G + S
Sbjct: 16 AWTAIIGGLAIHGKGREALDWFTQMQKAGINPNSITFTAILTACSHAGLTEEGKSLFESM 195
Query: 262 AKRCKVDSNPILATALVDMYSKCGCVESALTVFEGIADK-DAGAWNAMISGVAL 314
+ + + +VD+ + G ++ A E + K +A W A+++ L
Sbjct: 196 SSVYNIKPSMEHYGCMVDLMGRAGLLKEAREFIESMPVKPNAAIWGALLNACQL 357
Score = 31.2 bits (69), Expect = 1.2
Identities = 15/36 (41%), Positives = 22/36 (60%), Gaps = 1/36 (2%)
Frame = +1
Query: 176 MIDGYGKVGDVESARELFEEMPER-NAVSWSAMMAA 210
M+D G+ G ++ ARE E MP + NA W A++ A
Sbjct: 241 MVDLMGRAGLLKEAREFIESMPVKPNAAIWGALLNA 348
>CO979675
Length = 732
Score = 170 bits (431), Expect = 1e-42
Identities = 83/216 (38%), Positives = 134/216 (61%), Gaps = 3/216 (1%)
Frame = -2
Query: 300 KDAGAWNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGMVHQGLRL 359
+D WNAMISG+A+ GD +L++F +M G +P + TF+AV TAC++ GM H+GL+L
Sbjct: 731 RDIVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACSYSGMAHEGLQL 552
Query: 360 FEEMSCTYGVEPRGEHYACVVDLLSRAGMVKEAEKFMEKMGGLV---EADLNVWGAILNA 416
++MS Y +EP+ EHY C+VDLLSRAG+ EA + ++ + W A L+A
Sbjct: 551 LDKMSSLYEIEPKSEHYGCLVDLLSRAGLFGEAMVMIRRITSTSWNGSEETLAWRAFLSA 372
Query: 417 CRIYKNVDVGNRVWKKMIDKGVADCGTHILTYNMYREAGWDAEAKRVRSMVSEAGMKKKP 476
C + + R K+++ + G ++L N+Y +G ++A+RVR+M+ G+ K P
Sbjct: 371 CCNHGQAQLAERAAKRLL-RLENHSGVYVLLSNLYAASGKHSDARRVRNMMRNKGVDKAP 195
Query: 477 GCSIIEVGDEVEEFLAGDHSHPQAQELCKLLDSILM 512
GCS +E+ V EF+AG+ +HPQ +E+ +L+ + M
Sbjct: 194 GCSSVEIDGVVSEFIAGEETHPQMEEIHSVLEILHM 87
Score = 55.5 bits (132), Expect = 6e-08
Identities = 33/128 (25%), Positives = 59/128 (45%), Gaps = 7/128 (5%)
Frame = -2
Query: 199 RNAVSWSAMMAAYSRVSDFKQVLALFEEMQNRGMKPNDSVLVTVLTACAHLGALTQGL-W 257
R+ V W+AM++ + D L +F EM+ G+KP+D + V TAC++ G +GL
Sbjct: 731 RDIVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACSYSGMAHEGLQL 552
Query: 258 VHSYAKRCKVDSNPILATALVDMYSKCGCVESALTVFEGI------ADKDAGAWNAMISG 311
+ + +++ LVD+ S+ G A+ + I ++ AW A +S
Sbjct: 551 LDKMSSLYEIEPKSEHYGCLVDLLSRAGLFGEAMVMIRRITSTSWNGSEETLAWRAFLSA 372
Query: 312 VALSGDVR 319
G +
Sbjct: 371 CCNHGQAQ 348
>TC233590 similar to UP|Q6K9T2 (Q6K9T2) Pentatricopeptide repeat-containing
protein-like, partial (5%)
Length = 1094
Score = 164 bits (416), Expect = 7e-41
Identities = 96/300 (32%), Positives = 158/300 (52%), Gaps = 2/300 (0%)
Frame = +2
Query: 177 IDGYGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEMQNRGMKPND 236
+ GY GD+ES ELF +MP+ ++ SW++++ Y+R + L +F++M ++P+
Sbjct: 2 VSGYAVWGDMESGAELFSQMPKSDSCSWTSLIRGYARNGMGYEALGVFKQMIKHQVRPDQ 181
Query: 237 SVLVTVLTACAHLGALTQGLWVHSYAKRCKVDSNPILATALVDMYSKCGCVESALTVFEG 296
L T L ACA + +L G +H++ + N I+ A+V+MYSKCG +E+A VF
Sbjct: 182 FTLSTCLFACATIASLKHGRQIHAFLVLNNIKPNTIVVCAIVNMYSKCGSLETARRVFNF 361
Query: 297 IADK-DAGAWNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGMVHQ 355
I +K D WN MI +A G +++ + + M G +P + TFV +L AC H G+V +
Sbjct: 362 IGNKQDVVLWNTMILALAHYGYGIEAIMMLYNMLKIGVKPNKGTFVGILNACCHSGLVQE 541
Query: 356 GLRLFEEMSCTYGVEPRGEHYACVVDLLSRAGMVKEAEKFMEKMGGLVEADLNVWGAILN 415
GL+LF+ M+ +GV P EHY + +LL +A E+ K ++ M + +V + +
Sbjct: 542 GLQLFKSMTSEHGVVPDQEHYTRLANLLGQARCFNESVKDLQMMD--CKPGDHVCNSSIG 715
Query: 416 ACRIYKNVDVGNRVWKKMIDKGVADCGTHILTYNMYREAG-WDAEAKRVRSMVSEAGMKK 474
CR++ N+D G V +I + L Y G W+ K E G +K
Sbjct: 716 VCRMHGNIDHGAEVAAFLIKLQPQSSAAYELLSRTYAALGKWELVEKNKTYFG*EIGEEK 895
>TC223934
Length = 889
Score = 163 bits (413), Expect = 2e-40
Identities = 100/275 (36%), Positives = 145/275 (52%), Gaps = 31/275 (11%)
Frame = +2
Query: 39 LSAAAATSNFSYARSIFLHLTHRNTFIHNTMIRAYLLHAPSPAPALSCYSSMLRTGITVN 98
+SA + S + A S F ++ + N + N +IR +H AL Y MLR +
Sbjct: 83 ISACSNLSCINLAASAFANVQNPNVLVFNALIRG-CVHCCYSEQALVHYMHMLRNNVMPT 259
Query: 99 NYTFPPLIKASLALLSSSSSSNFTGRLVHGHVVKFGFHDDPYVVSGFIEFYCA------S 152
+Y+F LIKA L+ S+ G VHGHV K GF +V + IEFY S
Sbjct: 260 SYSFSSLIKACTLLVDSAF-----GEAVHGHVWKHGFDSHVFVQTTLIEFYSTFGDVGGS 424
Query: 153 RQ-------------------------LQTARLLFDQTTEKDVVLWTAMIDGYGKVGDVE 187
R+ + +A LFD+ EK+V W AMIDGYGK+G+ E
Sbjct: 425 RRVFDDMPERDVFAWTTMISAHVRDGDMASAGRLFDEMPEKNVATWNAMIDGYGKLGNAE 604
Query: 188 SARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEMQNRGMKPNDSVLVTVLTACA 247
SA LF +MP R+ +SW+ MM YSR +K+V+ALF ++ ++GM P++ + TV++ACA
Sbjct: 605 SAEFLFNQMPARDIISWTTMMNCYSRNKRYKEVIALFHDVIDKGMIPDEVTMTTVISACA 784
Query: 248 HLGALTQGLWVHSYAKRCKVDSNPILATALVDMYS 282
HLGAL G VH Y D + + ++L+DMY+
Sbjct: 785 HLGALALGKEVHLYLVLQGFDLDVYIGSSLIDMYA 889
Score = 59.7 bits (143), Expect = 3e-09
Identities = 39/163 (23%), Positives = 68/163 (40%)
Frame = +2
Query: 166 TEKDVVLWTAMIDGYGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFE 225
T +D L I + + A F + N + ++A++ +Q L +
Sbjct: 50 TTQDCFLVNQFISACSNLSCINLAASAFANVQNPNVLVFNALIRGCVHCCYSEQALVHYM 229
Query: 226 EMQNRGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAKRCKVDSNPILATALVDMYSKCG 285
M + P +++ AC L G VH + + DS+ + T L++ YS G
Sbjct: 230 HMLRNNVMPTSYSFSSLIKACTLLVDSAFGEAVHGHVWKHGFDSHVFVQTTLIEFYSTFG 409
Query: 286 CVESALTVFEGIADKDAGAWNAMISGVALSGDVRKSLELFHQM 328
V + VF+ + ++D AW MIS GD+ + LF +M
Sbjct: 410 DVGGSRRVFDDMPERDVFAWTTMISAHVRDGDMASAGRLFDEM 538
>AW757000 similar to PIR|C86440|C86 PPR-repeat protein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 442
Score = 139 bits (351), Expect = 2e-33
Identities = 58/137 (42%), Positives = 90/137 (65%)
Frame = +3
Query: 254 QGLWVHSYAKRCKVDSNPILATALVDMYSKCGCVESALTVFEGIADKDAGAWNAMISGVA 313
QG W+H+Y ++ + ++ TAL++MY+KCGC+E + +F G+ +KD +W ++I G+A
Sbjct: 6 QGKWIHNYIDENRIKVDAVVGTALIEMYAKCGCIEKSFEIFNGLKEKDTTSWTSIICGLA 185
Query: 314 LSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGMVHQGLRLFEEMSCTYGVEPRG 373
++G ++LELF M G +P + TFVAVL+AC+H G+V +G +LF MS Y +EP
Sbjct: 186 MNGKPSEALELFKAMQTCGLKPDDITFVAVLSACSHAGLVEEGRKLFHSMSSMYHIEPNL 365
Query: 374 EHYACVVDLLSRAGMVK 390
EHY C +DLL K
Sbjct: 366 EHYGCFIDLLGEPDFYK 416
Score = 69.7 bits (169), Expect = 3e-12
Identities = 34/103 (33%), Positives = 60/103 (58%), Gaps = 1/103 (0%)
Frame = +3
Query: 169 DVVLWTAMIDGYGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEMQ 228
D V+ TA+I+ Y K G +E + E+F + E++ SW++++ + + L LF+ MQ
Sbjct: 54 DAVVGTALIEMYAKCGCIEKSFEIFNGLKEKDTTSWTSIICGLAMNGKPSEALELFKAMQ 233
Query: 229 NRGMKPNDSVLVTVLTACAHLGALTQGLWV-HSYAKRCKVDSN 270
G+KP+D V VL+AC+H G + +G + HS + ++ N
Sbjct: 234 TCGLKPDDITFVAVLSACSHAGLVEEGRKLFHSMSSMYHIEPN 362
>TC224522 weakly similar to UP|Q9LUL5 (Q9LUL5) Emb|CAB66100.1, partial (18%)
Length = 534
Score = 129 bits (323), Expect = 4e-30
Identities = 58/165 (35%), Positives = 105/165 (63%)
Frame = +2
Query: 339 TFVAVLTACTHGGMVHQGLRLFEEMSCTYGVEPRGEHYACVVDLLSRAGMVKEAEKFMEK 398
TF+++L+AC G V++ + LF M YG+ PR EHYAC+VD++SRAG ++ A K + +
Sbjct: 11 TFLSLLSACCRAGKVNESMNLFSLMVDNYGIPPRSEHYACLVDVMSRAGQLQRACKIINE 190
Query: 399 MGGLVEADLNVWGAILNACRIYKNVDVGNRVWKKMIDKGVADCGTHILTYNMYREAGWDA 458
M +AD ++WGA+L AC ++ NV++G +++++ + G +++ N+Y AG
Sbjct: 191 MP--FKADSSIWGAVLAACSVHLNVELGELAARRILNLDPFNSGAYVMLSNIYAAAGKWK 364
Query: 459 EAKRVRSMVSEAGMKKKPGCSIIEVGDEVEEFLAGDHSHPQAQEL 503
+ R+R ++ E G+KK+ S +++G++ F+ GD SHP ++
Sbjct: 365 DVHRIRVLMKEQGVKKQTAYSWLQIGNKTHYFVGGDPSHPNINDI 499
>CK769031
Length = 827
Score = 79.7 bits (195), Expect(2) = 6e-30
Identities = 39/88 (44%), Positives = 58/88 (65%), Gaps = 1/88 (1%)
Frame = +2
Query: 311 GVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGMVHQGLRLFEEM-SCTYGV 369
G+A++G +++L+ F + G +P E TF+AV TAC H G+V +G + F EM S Y +
Sbjct: 14 GLAINGYGKEALKRFEDLVESGARPNEVTFLAVYTACCHNGLVDEGRKYFNEMTSPHYNL 193
Query: 370 EPRGEHYACVVDLLSRAGMVKEAEKFME 397
P EHY C+VDLL RAG+V EA + ++
Sbjct: 194 SPCLEHYGCMVDLLCRAGLVGEAVELIK 277
Score = 69.7 bits (169), Expect(2) = 6e-30
Identities = 39/115 (33%), Positives = 60/115 (51%)
Frame = +1
Query: 403 VEADLNVWGAILNACRIYKNVDVGNRVWKKMIDKGVADCGTHILTYNMYREAGWDAEAKR 462
V D + GA+L+AC NV + K + + D G ++L N+Y AE +
Sbjct: 268 VNKDFQIPGALLSACNT*GNVGFTQEMLKSLQNVEFQDSGIYVLLSNLYATNKKWAEVRS 447
Query: 463 VRSMVSEAGMKKKPGCSIIEVGDEVEEFLAGDHSHPQAQELCKLLDSILMMANLE 517
+R ++ + G+ K PG SII V EFL GD SHPQ++++ LL+ + LE
Sbjct: 448 IRRLMKQKGISKAPGSSIISVDGMSHEFLVGDISHPQSEDIYILLNILANQIYLE 612
Score = 37.4 bits (85), Expect = 0.016
Identities = 21/80 (26%), Positives = 38/80 (47%), Gaps = 2/80 (2%)
Frame = +2
Query: 218 KQVLALFEEMQNRGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAKRCKVDSNPILA--T 275
K+ L FE++ G +PN+ + V TAC H G + +G + + +P L
Sbjct: 38 KEALKRFEDLVESGARPNEVTFLAVYTACCHNGLVDEGRKYFNEMTSPHYNLSPCLEHYG 217
Query: 276 ALVDMYSKCGCVESALTVFE 295
+VD+ + G V A+ + +
Sbjct: 218 CMVDLLCRAGLVGEAVELIK 277
>TC232145
Length = 817
Score = 128 bits (321), Expect = 7e-30
Identities = 68/179 (37%), Positives = 104/179 (57%)
Frame = +3
Query: 332 GTQPTETTFVAVLTACTHGGMVHQGLRLFEEMSCTYGVEPRGEHYACVVDLLSRAGMVKE 391
G P TFV+VL+AC+H G + G + F M+ + ++P EHYAC+VDLL R G ++E
Sbjct: 6 GVVPEYITFVSVLSACSHTGKIDDGFKYFNSMANVHNIKPGLEHYACMVDLLGRVGRLEE 185
Query: 392 AEKFMEKMGGLVEADLNVWGAILNACRIYKNVDVGNRVWKKMIDKGVADCGTHILTYNMY 451
A +F+E M E D VWGA+L AC + NV++G V +++ + G ++L N+Y
Sbjct: 186 ACRFIESMP--FEPDSLVWGALLGACGKHANVEMGREVAERLFKLEPDNPGNYMLLSNIY 359
Query: 452 REAGWDAEAKRVRSMVSEAGMKKKPGCSIIEVGDEVEEFLAGDHSHPQAQELCKLLDSI 510
G EA VR ++ G++K+ GCS I+V + F A D SH + QE+ +L +
Sbjct: 360 IRHGMLEEADEVRRLMGINGVRKESGCSWIDVKNRTFVFNANDRSHSRTQEIYGMLQKL 536
Score = 35.4 bits (80), Expect = 0.062
Identities = 23/100 (23%), Positives = 44/100 (44%), Gaps = 2/100 (2%)
Frame = +3
Query: 231 GMKPNDSVLVTVLTACAHLGALTQGL-WVHSYAKRCKVDSNPILATALVDMYSKCGCVES 289
G+ P V+VL+AC+H G + G + +S A + +VD+ + G +E
Sbjct: 6 GVVPEYITFVSVLSACSHTGKIDDGFKYFNSMANVHNIKPGLEHYACMVDLLGRVGRLEE 185
Query: 290 ALTVFEGIA-DKDAGAWNAMISGVALSGDVRKSLELFHQM 328
A E + + D+ W A++ +V E+ ++
Sbjct: 186 ACRFIESMPFEPDSLVWGALLGACGKHANVEMGREVAERL 305
>AW396441
Length = 459
Score = 127 bits (320), Expect = 9e-30
Identities = 60/150 (40%), Positives = 90/150 (60%)
Frame = +3
Query: 242 VLTACAHLGALTQGLWVHSYAKRCKVDSNPILATALVDMYSKCGCVESALTVFEGIADKD 301
V+ A +++ +L G H+ + +D +P + +LVDMY+KCG +E + F +D
Sbjct: 12 VIAAASNIASLRHGQQFHNQVIKMGLDDDPFVTNSLVDMYAKCGSIEESHKAFSSTNQRD 191
Query: 302 AGAWNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGMVHQGLRLFE 361
WN+MIS A GD K+LE+F +M G +P TFV +L+AC+H G++ G FE
Sbjct: 192 IACWNSMISTYAQHGDAAKALEVFERMIMEGVKPNYVTFVGLLSACSHAGLLDLGFHHFE 371
Query: 362 EMSCTYGVEPRGEHYACVVDLLSRAGMVKE 391
MS +G+EP +HYAC+V LL RAG + E
Sbjct: 372 SMS-KFGIEPXIDHYACMVSLLGRAGKIYE 458
Score = 70.5 bits (171), Expect = 2e-12
Identities = 42/166 (25%), Positives = 73/166 (43%)
Frame = +3
Query: 108 ASLALLSSSSSSNFTGRLVHGHVVKFGFHDDPYVVSGFIEFYCASRQLQTARLLFDQTTE 167
A++ +S+ +S G+ H V+K G DDP+V +
Sbjct: 6 AAVIAAASNIASLRHGQQFHNQVIKMGLDDDPFVTN------------------------ 113
Query: 168 KDVVLWTAMIDGYGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEM 227
+++D Y K G +E + + F +R+ W++M++ Y++ D + L +FE M
Sbjct: 114 -------SLVDMYAKCGSIEESHKAFSSTNQRDIACWNSMISTYAQHGDAAKALEVFERM 272
Query: 228 QNRGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAKRCKVDSNPIL 273
G+KPN V +L+AC+H G L G H + K P +
Sbjct: 273 IMEGVKPNYVTFVGLLSACSHAGLLDLGF--HHFESMSKFGIEPXI 404
>AW234248
Length = 399
Score = 122 bits (307), Expect = 3e-28
Identities = 56/125 (44%), Positives = 80/125 (63%)
Frame = +2
Query: 275 TALVDMYSKCGCVESALTVFEGIADKDAGAWNAMISGVALSGDVRKSLELFHQMAAHGTQ 334
T+LVDMY+KCG +E A +F+ +WNAMI G+A G+ ++L+ F +M + G
Sbjct: 8 TSLVDMYAKCGNIEDARGLFKRTNTSRIASWNAMIVGLAQHGNAEEALQFFEEMKSRGVT 187
Query: 335 PTETTFVAVLTACTHGGMVHQGLRLFEEMSCTYGVEPRGEHYACVVDLLSRAGMVKEAEK 394
P TF+ VL+AC+H G+V + F M YG+EP EHY+C+VD LSRAG ++EAEK
Sbjct: 188 PDRVTFIGVLSACSHSGLVSEAYENFYSMQKIYGIEPEIEHYSCLVDALSRAGRIREAEK 367
Query: 395 FMEKM 399
+ M
Sbjct: 368 VISSM 382
Score = 68.6 bits (166), Expect = 7e-12
Identities = 37/125 (29%), Positives = 67/125 (53%), Gaps = 1/125 (0%)
Frame = +2
Query: 174 TAMIDGYGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEMQNRGMK 233
T+++D Y K G++E AR LF+ SW+AM+ ++ + ++ L FEEM++RG+
Sbjct: 8 TSLVDMYAKCGNIEDARGLFKRTNTSRIASWNAMIVGLAQHGNAEEALQFFEEMKSRGVT 187
Query: 234 PNDSVLVTVLTACAHLGALTQGLW-VHSYAKRCKVDSNPILATALVDMYSKCGCVESALT 292
P+ + VL+AC+H G +++ +S K ++ + LVD S+ G + A
Sbjct: 188 PDRVTFIGVLSACSHSGLVSEAYENFYSMQKIYGIEPEIEHYSCLVDALSRAGRIREAEK 367
Query: 293 VFEGI 297
V +
Sbjct: 368 VISSM 382
>TC234029 similar to UP|Q9LN01 (Q9LN01) T6D22.15, partial (32%)
Length = 732
Score = 119 bits (297), Expect = 4e-27
Identities = 58/144 (40%), Positives = 93/144 (64%), Gaps = 1/144 (0%)
Frame = +2
Query: 368 GVEPRGEHYACVVDLLSRAGMVKEAEKFMEKMGGLVEADLNVWGAILNACRIYKNVDVGN 427
G+ P+ +HY C++DLL+R+G EA+ M M +E D +WG++LNACRI+ V+ G
Sbjct: 2 GISPKLQHYGCMIDLLARSGKFDEAKVLMGNME--MEPDGAIWGSLLNACRIHGQVEFGE 175
Query: 428 RVWKKMIDKGVADCGTHILTYNMYREAG-WDAEAKRVRSMVSEAGMKKKPGCSIIEVGDE 486
V +++ + + G ++L N+Y AG WD AK +R+ +++ GMKK PGC+ IE+
Sbjct: 176 YVAERLFELEPENSGAYVLLSNIYAGAGRWDDVAK-IRTKLNDKGMKKVPGCTSIEIDGV 352
Query: 487 VEEFLAGDHSHPQAQELCKLLDSI 510
V EFL GD HPQ++ + ++LD +
Sbjct: 353 VHEFLVGDKFHPQSENIFRMLDEV 424
>BI893521
Length = 423
Score = 117 bits (294), Expect = 9e-27
Identities = 55/142 (38%), Positives = 88/142 (61%)
Frame = +1
Query: 293 VFEGIADKDAGAWNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGM 352
VF + ++ W ++ISG A G K+LELF++M G +P E T++AVL+AC+H G+
Sbjct: 4 VFNDMGYRNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSACSHVGL 183
Query: 353 VHQGLRLFEEMSCTYGVEPRGEHYACVVDLLSRAGMVKEAEKFMEKMGGLVEADLNVWGA 412
+ + + F M + + PR EHYAC+VDLL R+G++ EA +F+ M +AD VW
Sbjct: 184 IDEAWKHFNSMHYNHSISPRMEHYACMVDLLGRSGLLLEAIEFINSMP--FDADALVWRT 357
Query: 413 ILNACRIYKNVDVGNRVWKKMI 434
L +CR+++N +G KK++
Sbjct: 358 FLGSCRVHRNTKLGEHAAKKIL 423
Score = 60.1 bits (144), Expect = 2e-09
Identities = 31/122 (25%), Positives = 63/122 (51%), Gaps = 3/122 (2%)
Frame = +1
Query: 191 ELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEMQNRGMKPNDSVLVTVLTACAHLG 250
++F +M RN ++W+++++ +++ + L LF EM G+KPN+ + VL+AC+H+G
Sbjct: 1 QVFNDMGYRNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSACSHVG 180
Query: 251 ALTQGLWVHSYAKRCKVDSNPILA--TALVDMYSKCGCVESALTVFEGIA-DKDAGAWNA 307
+ + W H + +P + +VD+ + G + A+ + D DA W
Sbjct: 181 LIDEA-WKHFNSMHYNHSISPRMEHYACMVDLLGRSGLLLEAIEFINSMPFDADALVWRT 357
Query: 308 MI 309
+
Sbjct: 358 FL 363
>AW317686
Length = 447
Score = 112 bits (281), Expect = 3e-25
Identities = 51/143 (35%), Positives = 86/143 (59%)
Frame = +1
Query: 293 VFEGIADKDAGAWNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGM 352
+FE +D WN +I+G A +G +SL +F +M +P TF+ VL+ C H G+
Sbjct: 7 LFEMAPMRDVVTWNTLITGFAQNGHGEESLAVFRRMIEAKVEPNHVTFLGVLSGCNHAGL 186
Query: 353 VHQGLRLFEEMSCTYGVEPRGEHYACVVDLLSRAGMVKEAEKFMEKMGGLVEADLNVWGA 412
++GL+L + M YGV+P+ EHYA ++DLL R + EA +EK+ ++ + VWGA
Sbjct: 187 DNEGLQLVDLMERQYGVKPKAEHYALLIDLLGRRNRLMEAMSLIEKVPDGIKNHIAVWGA 366
Query: 413 ILNACRIYKNVDVGNRVWKKMID 435
+L AC ++ N+D+ + +K+ +
Sbjct: 367 VLGACXVHGNLDLARKAAEKLFE 435
Score = 50.4 bits (119), Expect = 2e-06
Identities = 33/148 (22%), Positives = 69/148 (46%), Gaps = 11/148 (7%)
Frame = +1
Query: 192 LFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEMQNRGMKPNDSVLVTVLTACAHLGA 251
LFE P R+ V+W+ ++ +++ ++ LA+F M ++PN + VL+ C H G
Sbjct: 7 LFEMAPMRDVVTWNTLITGFAQNGHGEESLAVFRRMIEAKVEPNHVTFLGVLSGCNHAGL 186
Query: 252 LTQGLWV-----HSYAKRCKVDSNPILATALVDMYSKCGCVESALTVFEGIAD---KDAG 303
+GL + Y + K + + L+D+ + + A+++ E + D
Sbjct: 187 DNEGLQLVDLMERQYGVKPKAEHYAL----LIDLLGRRNRLMEAMSLIEKVPDGIKNHIA 354
Query: 304 AWNAMISGVALSGDV---RKSLELFHQM 328
W A++ + G++ RK+ E ++
Sbjct: 355 VWGAVLGACXVHGNLDLARKAAEKLFEL 438
>TC221986 weakly similar to GB|BAC16401.1|23237826|AP003749 selenium-binding
protein-like {Oryza sativa (japonica cultivar-group);} ,
partial (10%)
Length = 853
Score = 109 bits (273), Expect = 3e-24
Identities = 68/220 (30%), Positives = 114/220 (50%)
Frame = +1
Query: 245 ACAHLGALTQGLWVHSYAKRCKVDSNPILATALVDMYSKCGCVESALTVFEGIADKDAGA 304
AC +G+L G VH +A + + P L TAL+DMYSKCG ++ A TVF+ + +
Sbjct: 4 ACTEMGSLKLGRRVHDFALKNGFELEPFLGTALIDMYSKCGNLDDARTVFDMMQMRTLAT 183
Query: 305 WNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGMVHQGLRLFEEMS 364
WN MI+ + + G ++L +F +M P TFV VL+AC + + + F M+
Sbjct: 184 WNTMITSLGVHGYRDEALSIFEEMEKANEVPDAITFVGVLSACVYMNDLELAQKYFNLMT 363
Query: 365 CTYGVEPRGEHYACVVDLLSRAGMVKEAEKFMEKMGGLVEADLNVWGAILNACRIYKNVD 424
YG+ P EHY C+V++ +RA +K E +M G +EA+ +V +L+ ++ D
Sbjct: 364 DHYGITPILEHYTCMVEIHTRA--IKLDEIYMS--GNTMEANHDV-AELLHKNKLTSFDD 528
Query: 425 VGNRVWKKMIDKGVADCGTHILTYNMYREAGWDAEAKRVR 464
+ + KK G D +L ++ +G + ++R
Sbjct: 529 IKKLIHKKY---GDLDFSELVLDHSSTSSSGMQIDISKIR 639
Score = 55.1 bits (131), Expect = 8e-08
Identities = 28/86 (32%), Positives = 47/86 (54%)
Frame = +1
Query: 167 EKDVVLWTAMIDGYGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEE 226
E + L TA+ID Y K G+++ AR +F+ M R +W+ M+ + + L++FEE
Sbjct: 73 ELEPFLGTALIDMYSKCGNLDDARTVFDMMQMRTLATWNTMITSLGVHGYRDEALSIFEE 252
Query: 227 MQNRGMKPNDSVLVTVLTACAHLGAL 252
M+ P+ V VL+AC ++ L
Sbjct: 253 MEKANEVPDAITFVGVLSACVYMNDL 330
Score = 53.5 bits (127), Expect = 2e-07
Identities = 33/117 (28%), Positives = 56/117 (47%), Gaps = 5/117 (4%)
Frame = +1
Query: 123 GRLVHGHVVKFGFHDDPYVVSGFIEFYCASRQLQTARLLFDQTTEKDVVLWTAMIDGYGK 182
GR VH +K GF +P++ + I+ Y L AR +FD + + W MI G
Sbjct: 34 GRRVHDFALKNGFELEPFLGTALIDMYSKCGNLDDARTVFDMMQMRTLATWNTMITSLGV 213
Query: 183 VGDVESARELFEEMPERN----AVSWSAMMAAYSRVSDFKQVLALFEEMQNR-GMKP 234
G + A +FEEM + N A+++ +++A ++D + F M + G+ P
Sbjct: 214 HGYRDEALSIFEEMEKANEVPDAITFVGVLSACVYMNDLELAQKYFNLMTDHYGITP 384
>BE802290
Length = 481
Score = 108 bits (271), Expect = 4e-24
Identities = 55/157 (35%), Positives = 86/157 (54%)
Frame = -1
Query: 325 FHQMAAHGTQPTETTFVAVLTACTHGGMVHQGLRLFEEMSCTYGVEPRGEHYACVVDLLS 384
F Q ++P F+A+ AC+H + +G FE MS Y ++P EHY C+VDL+
Sbjct: 481 FTQCRKQKSKPNSIPFMAISPACSHAALTEEGTSSFESMSSVYNIKPSMEHYGCMVDLMG 302
Query: 385 RAGMVKEAEKFMEKMGGLVEADLNVWGAILNACRIYKNVDVGNRVWKKMIDKGVADCGTH 444
AG++KEA +F+E M V+ + +WGA+LNAC+++K ++G K I+ G +
Sbjct: 301 GAGLLKEAREFVESMP--VKPNAAIWGALLNACQLHKYFELGKETGKIQIELDPDHSGRY 128
Query: 445 ILTYNMYREAGWDAEAKRVRSMVSEAGMKKKPGCSII 481
I ++Y A + RVRS + G+ GCS I
Sbjct: 127 IHLASIYAAAREWNQVTRVRSQIKHRGLLNYSGCSSI 17
Score = 29.3 bits (64), Expect = 4.4
Identities = 15/36 (41%), Positives = 21/36 (57%), Gaps = 1/36 (2%)
Frame = -1
Query: 176 MIDGYGKVGDVESARELFEEMPER-NAVSWSAMMAA 210
M+D G G ++ ARE E MP + NA W A++ A
Sbjct: 319 MVDLMGGAGLLKEAREFVESMPVKPNAAIWGALLNA 212
>TC234676
Length = 914
Score = 105 bits (262), Expect = 5e-23
Identities = 63/227 (27%), Positives = 110/227 (47%)
Frame = +1
Query: 169 DVVLWTAMIDGYGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEMQ 228
+V + ++++ Y K G VE +R LF+ M +RN +SW+AM+ +Y + L + MQ
Sbjct: 58 NVSVASSLMTMYSKCGVVEYSRRLFDNMEQRNVISWTAMIDSYIENGYLCEALGVIRSMQ 237
Query: 229 NRGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAKRCKVDSNPILATALVDMYSKCGCVE 288
+P+ + +L+ C + G +H + S ++ L++MY G +
Sbjct: 238 LSKHRPDSVAIGRMLSVCGERKLVKLGKEIHGQILKRDFTSVHFVSAELINMYGFFGDIN 417
Query: 289 SALTVFEGIADKDAGAWNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACT 348
A VF + K + W A+I + + ++ LF QM P TF A+L+ C
Sbjct: 418 KANLVFNAVPVKGSMTWTALIRAYGYNELYQDAVNLFDQMR---YSPNHFTFEAILSICD 588
Query: 349 HGGMVHQGLRLFEEMSCTYGVEPRGEHYACVVDLLSRAGMVKEAEKF 395
G V R+F M Y +E EH+A +V LL+ G +++A++F
Sbjct: 589 KAGFVDDACRIFNSMP-RYKIEASKEHFAIMVRLLTHNGQLEKAQRF 726
Score = 52.8 bits (125), Expect = 4e-07
Identities = 28/103 (27%), Positives = 53/103 (51%)
Frame = +1
Query: 254 QGLWVHSYAKRCKVDSNPILATALVDMYSKCGCVESALTVFEGIADKDAGAWNAMISGVA 313
+G+ +H+YA + N +A++L+ MYSKCG VE + +F+ + ++ +W AMI
Sbjct: 10 KGMQIHAYALKHWFLPNVSVASSLMTMYSKCGVVEYSRRLFDNMEQRNVISWTAMIDSYI 189
Query: 314 LSGDVRKSLELFHQMAAHGTQPTETTFVAVLTACTHGGMVHQG 356
+G + ++L + M +P +L+ C +V G
Sbjct: 190 ENGYLCEALGVIRSMQLSKHRPDSVAIGRMLSVCGERKLVKLG 318
>BQ273653
Length = 376
Score = 103 bits (257), Expect = 2e-22
Identities = 54/125 (43%), Positives = 77/125 (61%), Gaps = 1/125 (0%)
Frame = +2
Query: 162 FDQTTEKDVVLWTAMIDGYGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVL 221
FD+ E+D V W MIDGY K+GDV +AR LF+EMP R+ +S ++MMA Y + + L
Sbjct: 2 FDEMPERDSVSWVTMIDGYVKLGDVLAARRLFDEMPSRDVISCNSMMAGYVQNGCCIEAL 181
Query: 222 ALFEEMQN-RGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAKRCKVDSNPILATALVDM 280
+F +M+ + P+D+ L+ V+TA A LG + G+ +H Y N L AL+DM
Sbjct: 182 KIFYDMKRATNVVPDDTTLLIVITAFAQLGHVEDGVVIHHYLMDKGYSLNGKLGVALIDM 361
Query: 281 YSKCG 285
YSKCG
Sbjct: 362 YSKCG 376
>AW185734
Length = 337
Score = 101 bits (251), Expect = 9e-22
Identities = 46/111 (41%), Positives = 73/111 (65%)
Frame = +3
Query: 184 GDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEMQNRGMKPNDSVLVTVL 243
G++ +ARELF+ M +R+ +SW+ M+ +YS+ F + L LF+EM +KP++ + +VL
Sbjct: 3 GNLVAARELFDAMSQRDVISWTNMITSYSQAGQFTEALKLFKEMMESKVKPDEITVASVL 182
Query: 244 TACAHLGALTQGLWVHSYAKRCKVDSNPILATALVDMYSKCGCVESALTVF 294
+ACAH G+L G H Y ++ V ++ + AL+DMY +CG VE AL VF
Sbjct: 183 SACAHTGSLDVGEAAHDYIQKYDVKADIYVGNALIDMYCRCGVVEKALEVF 335
>BM177962
Length = 421
Score = 101 bits (251), Expect = 9e-22
Identities = 51/137 (37%), Positives = 79/137 (57%)
Frame = +2
Query: 227 MQNRGMKPNDSVLVTVLTACAHLGALTQGLWVHSYAKRCKVDSNPILATALVDMYSKCGC 286
++ R PN +V VL ACA L AL QG +H Y R +DS + AL+ MY +CG
Sbjct: 2 LEARDSVPNSVTMVNVLQACAGLAALEQGKLIHGYILRRGLDSILPVLNALITMYGRCGE 181
Query: 287 VESALTVFEGIADKDAGAWNAMISGVALSGDVRKSLELFHQMAAHGTQPTETTFVAVLTA 346
+ VF+ + ++D +WN++IS + G +K++++F M G P+ +F+ VL A
Sbjct: 182 ILMGQRVFDNMKNRDVVSWNSLISIYGMHGFGKKAIQIF*NMIHQGGSPSYISFITVLGA 361
Query: 347 CTHGGMVHQGLRLFEEM 363
C+H G+V +G LFE M
Sbjct: 362 CSHAGLVEEGKILFESM 412
Score = 56.6 bits (135), Expect = 3e-08
Identities = 25/81 (30%), Positives = 48/81 (58%)
Frame = +2
Query: 175 AMIDGYGKVGDVESARELFEEMPERNAVSWSAMMAAYSRVSDFKQVLALFEEMQNRGMKP 234
A+I YG+ G++ + +F+ M R+ VSW+++++ Y K+ + +F M ++G P
Sbjct: 149 ALITMYGRCGEILMGQRVFDNMKNRDVVSWNSLISIYGMHGFGKKAIQIF*NMIHQGGSP 328
Query: 235 NDSVLVTVLTACAHLGALTQG 255
+ +TVL AC+H G + +G
Sbjct: 329 SYISFITVLGACSHAGLVEEG 391
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.133 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,083,094
Number of Sequences: 63676
Number of extensions: 355818
Number of successful extensions: 1969
Number of sequences better than 10.0: 146
Number of HSP's better than 10.0 without gapping: 1799
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1915
length of query: 519
length of database: 12,639,632
effective HSP length: 102
effective length of query: 417
effective length of database: 6,144,680
effective search space: 2562331560
effective search space used: 2562331560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0252d.2


