

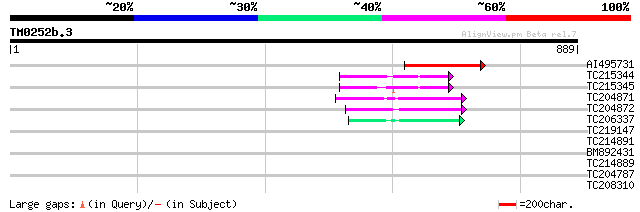
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252b.3
(889 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AI495731 weakly similar to GP|23429518|gb APC2 {Arabidopsis thal... 223 3e-58
TC215344 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, p... 62 8e-10
TC215345 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, p... 62 1e-09
TC204871 similar to UP|Q84LL5 (Q84LL5) Cullin 4, partial (81%) 59 7e-09
TC204872 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (42%) 57 3e-08
TC206337 similar to UP|Q7Y1Y8 (Q7Y1Y8) Cullin 3a (Fragment), par... 50 3e-06
TC219147 similar to PIR|T01092|T01092 cullin-like protein T10P11... 30 3.6
TC214891 homologue to PIR|C86216|C86216 protein T23G18.6 [import... 30 4.7
BM892431 similar to GP|20453225|gb| At5g26752 {Arabidopsis thali... 30 4.7
TC214889 homologue to PIR|C86216|C86216 protein T23G18.6 [import... 30 4.7
TC204787 similar to UP|Q84WK9 (Q84WK9) At4g33985, partial (61%) 30 6.1
TC208310 weakly similar to UP|Q9MAL3 (Q9MAL3) F27F5.3, partial (... 29 8.0
>AI495731 weakly similar to GP|23429518|gb APC2 {Arabidopsis thaliana},
partial (14%)
Length = 386
Score = 223 bits (568), Expect = 3e-58
Identities = 104/126 (82%), Positives = 116/126 (91%)
Frame = +1
Query: 620 WPPIQVEPLNLPEPVDKLLSDYAKRFNEIKTPRKLQWKKSLGTVKLELQLKDRVLQFTVA 679
WPPIQ EPLNLPEPVD+LLSDYAKRFNEIKTPRKLQWKKSLGT+KLELQ +DR +QFTVA
Sbjct: 7 WPPIQDEPLNLPEPVDQLLSDYAKRFNEIKTPRKLQWKKSLGTIKLELQFQDREIQFTVA 186
Query: 680 PVHASIIMNFQDQTSWTSKNLAAAVGIPVDALNRRMSFWISKGVVAESSGGDSSDHVYTI 739
PVHASIIM FQDQ +WTSKNLAAA+GIP D LNRR++FWISKG++AES G DSSDHVYTI
Sbjct: 187 PVHASIIMKFQDQPNWTSKNLAAAIGIPADVLNRRINFWISKGIIAESQGADSSDHVYTI 366
Query: 740 MESMVE 745
+E+M E
Sbjct: 367 VENMAE 384
>TC215344 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, partial (92%)
Length = 2465
Score = 62.4 bits (150), Expect = 8e-10
Identities = 50/183 (27%), Positives = 82/183 (44%), Gaps = 3/183 (1%)
Frame = +1
Query: 517 IVGIIGSKDQLVHEYRTMLAEKLLNKSDYDIDSEIRTLELLKIHFGESSLQKCEIMLNDL 576
++ I KD YR LA +LL + D E L LK G K E M+ DL
Sbjct: 1093 LLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 1272
Query: 577 IGSKRINSNIKATISQPSQTNVEVEDNAISMDNIAATIISSNFWPPIQVEPLNLPEPVDK 636
+K ++ + +S NA ++ T++++ FWP + LNLP + +
Sbjct: 1273 TLAKENQTSFEEYLSN--------NPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMIR 1428
Query: 637 LLSDYAKRFNEIKTP-RKLQWKKSLGTVKLELQLKDRVLQFTVAPVHAS--IIMNFQDQT 693
+ + K F + KT RKL W SLGT + + + ++ V AS ++ N D+
Sbjct: 1429 CVEVF-KEFYQTKTKHRKLTWIYSLGTCNISGKFDPKTVELIVTTYQASALLLFNSSDRL 1605
Query: 694 SWT 696
S++
Sbjct: 1606 SYS 1614
>TC215345 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, partial (77%)
Length = 2106
Score = 62.0 bits (149), Expect = 1e-09
Identities = 52/187 (27%), Positives = 84/187 (44%), Gaps = 7/187 (3%)
Frame = +1
Query: 517 IVGIIGSKDQLVHEYRTMLAEKLLNKSDYDIDSEIRTLELLKIHFGESSLQKCEIMLNDL 576
++ I KD YR LA +LL + D E L LK G K E M+ DL
Sbjct: 766 LLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 945
Query: 577 IGSKRINSNIKATISQPSQTNVEV----EDNAISMDNIAATIISSNFWPPIQVEPLNLPE 632
T+++ +QT+ E NA ++ T++++ FWP + LNLP
Sbjct: 946 ------------TLAKENQTSFEEYLTNNPNADPGIDLTVTVLTTGFWPSYKSFDLNLPA 1089
Query: 633 PVDKLLSDYAKRFNEIKTP-RKLQWKKSLGTVKLELQLKDRVLQFTVAPVHAS--IIMNF 689
+ + + + K F + KT RKL W SLGT + + + ++ V AS ++ N
Sbjct: 1090EMVRCVEVF-KEFYQTKTKHRKLTWIYSLGTCNISGKFDPKTVELIVTTYQASALLLFNS 1266
Query: 690 QDQTSWT 696
D+ S++
Sbjct: 1267SDRLSYS 1287
>TC204871 similar to UP|Q84LL5 (Q84LL5) Cullin 4, partial (81%)
Length = 2087
Score = 59.3 bits (142), Expect = 7e-09
Identities = 50/206 (24%), Positives = 89/206 (42%), Gaps = 1/206 (0%)
Frame = +1
Query: 511 VDILGMIVGIIGSKDQLVHEYRTMLAEKLLNKSDYDIDSEIRTLELLKIHFGESSLQKCE 570
+D + ++ I KD Y+ LA++LL ID+E + LK G K E
Sbjct: 829 LDKVLVLFRFIQGKDVFEAFYKKDLAKRLLLGKSASIDAEKSMISKLKTECGSQFTNKLE 1008
Query: 571 IMLNDLIGSKRINSNIKATISQPSQTNVEVEDNAISMDNIAATIISSNFWPPIQVEPLNL 630
M D+ SK IN + K Q SQ ++ ++ ++++ +WP + L
Sbjct: 1009 GMFKDIELSKEINESFK----QSSQARTKLPSGI----EMSVHVLTTGYWPTYPPMDVRL 1164
Query: 631 PEPVDKLLSDYAKRFNEIK-TPRKLQWKKSLGTVKLELQLKDRVLQFTVAPVHASIIMNF 689
P ++ + D K F K + R+L W+ SLG L+ + + V+ ++M F
Sbjct: 1165 PHELN-VYQDIFKEFYLSKYSGRRLMWQNSLGHCVLKAEFPKGKKELAVSLFQTVVLMLF 1341
Query: 690 QDQTSWTSKNLAAAVGIPVDALNRRM 715
D + +++ + GI L R +
Sbjct: 1342 NDAEKLSFQDIKDSTGIEDKELRRTL 1419
>TC204872 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (42%)
Length = 1224
Score = 57.4 bits (137), Expect = 3e-08
Identities = 47/190 (24%), Positives = 80/190 (41%), Gaps = 1/190 (0%)
Frame = +2
Query: 527 LVHEYRTMLAEKLLNKSDYDIDSEIRTLELLKIHFGESSLQKCEIMLNDLIGSKRINSNI 586
L H R +L +LL ID+E + LK G K E M D+ SK IN +
Sbjct: 2 LKHSIRKILLRRLLLGKSASIDAEKSMISKLKTECGSQFTNKLEGMFKDIELSKEINDSF 181
Query: 587 KATISQPSQTNVEVEDNAISMDNIAATIISSNFWPPIQVEPLNLPEPVDKLLSDYAKRFN 646
K + S+ +E ++ ++++ WP + LP ++ + D K F
Sbjct: 182 KQSSQARSKLASGIE--------MSVHVLTTGHWPTYPPMDVRLPHELN-VYQDIFKEFY 334
Query: 647 EIK-TPRKLQWKKSLGTVKLELQLKDRVLQFTVAPVHASIIMNFQDQTSWTSKNLAAAVG 705
K + R+L W+ SLG L+ + + V+ ++M F D + +++ A G
Sbjct: 335 LSKYSGRRLMWQNSLGHCVLKAEFPKGRKELAVSLFQTVVLMLFNDAEKLSLQDIKDATG 514
Query: 706 IPVDALNRRM 715
I L R +
Sbjct: 515 IEDKELRRTL 544
>TC206337 similar to UP|Q7Y1Y8 (Q7Y1Y8) Cullin 3a (Fragment), partial (91%)
Length = 1135
Score = 50.4 bits (119), Expect = 3e-06
Identities = 41/184 (22%), Positives = 73/184 (39%), Gaps = 1/184 (0%)
Frame = +3
Query: 531 YRTMLAEKLLNKSDYDIDSEIRTLELLKIHFGESSLQKCEIMLNDLIGSKRINSNIKATI 590
Y+ LA++LL+ D+E + LK G K E M D+ S A +
Sbjct: 9 YKQHLAKRLLSGKTISDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSHDTMQGFYAIL 188
Query: 591 SQPSQTNVEVEDNAISMDNIAATIISSNFWPPIQVEPLNLPEPVDKLLSDYAKRFNEIKT 650
E+ D + ++ ++++ WP P NLP + + + +
Sbjct: 189 G------TELGDGPL----LSVQVLTTGSWPTQPSPPCNLPVEILGVCDKFRTYYLGTHN 338
Query: 651 PRKLQWKKSLGTVKLELQL-KDRVLQFTVAPVHASIIMNFQDQTSWTSKNLAAAVGIPVD 709
R+L W+ ++GT L+ K + + V+ ++M F T K + A IP+
Sbjct: 339 GRRLSWQTNMGTADLKATFGKGQKHELNVSTYQMCVLMLFNSAERLTCKEIEQATAIPMS 518
Query: 710 ALNR 713
L R
Sbjct: 519 DLRR 530
>TC219147 similar to PIR|T01092|T01092 cullin-like protein T10P11.14.1 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(33%)
Length = 1237
Score = 30.4 bits (67), Expect = 3.6
Identities = 23/72 (31%), Positives = 38/72 (51%), Gaps = 3/72 (4%)
Frame = +3
Query: 628 LNLPEPVDKLLSDYAKRFNEIKTP-RKLQWKKSLGTVKLELQLKDRVLQFTVA--PVHAS 684
LNLP + + L + K F E +T RKL W SLGT + + + + ++ V P A
Sbjct: 18 LNLPSEMIRCLEVF-KGFYETRTKHRKLTWIYSLGTCHVTGKFETKNIELIVPTYPAAAL 194
Query: 685 IIMNFQDQTSWT 696
++ N D+ S++
Sbjct: 195 LLFNNADRLSYS 230
>TC214891 homologue to PIR|C86216|C86216 protein T23G18.6 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(66%)
Length = 1257
Score = 30.0 bits (66), Expect = 4.7
Identities = 27/80 (33%), Positives = 34/80 (41%), Gaps = 2/80 (2%)
Frame = +3
Query: 278 LLKAKVYNVAGDDFRGSILQSI--QRWIQAVPLQFLHALLVYIGDSVSYESTSSGLKSPL 335
L K Y V +D I SI QRW A Q + L+ G E T + +
Sbjct: 75 LRKDPAYYVLKEDESPCIFGSIEKQRWSYACAKQLIERLIYAEGAENGLEFTIVRPFNWI 254
Query: 336 APQPSSFCSGIDTPSEGLVR 355
P+ F GID PSEG+ R
Sbjct: 255 GPR-MDFIPGIDGPSEGVPR 311
>BM892431 similar to GP|20453225|gb| At5g26752 {Arabidopsis thaliana},
partial (9%)
Length = 421
Score = 30.0 bits (66), Expect = 4.7
Identities = 15/48 (31%), Positives = 24/48 (49%)
Frame = -3
Query: 456 DSLLEELNRDEEIQENVGVDDDFNTDDREAWINASRWQPDPVEADPLK 503
DS E E+ + G D + EAW N+S+ +P P++A L+
Sbjct: 317 DSTRREGKSSLEMSSSSG*TDSAAESESEAWSNSSKSRPSPLKASSLR 174
>TC214889 homologue to PIR|C86216|C86216 protein T23G18.6 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(97%)
Length = 1671
Score = 30.0 bits (66), Expect = 4.7
Identities = 27/80 (33%), Positives = 34/80 (41%), Gaps = 2/80 (2%)
Frame = +3
Query: 278 LLKAKVYNVAGDDFRGSILQSI--QRWIQAVPLQFLHALLVYIGDSVSYESTSSGLKSPL 335
L K Y V +D I SI QRW A Q + L+ G E T + +
Sbjct: 528 LRKDPAYYVLKEDESPCIFGSIEKQRWSYACAKQLIERLIFAEGAENGLEFTIVRPFNWI 707
Query: 336 APQPSSFCSGIDTPSEGLVR 355
P+ F GID PSEG+ R
Sbjct: 708 GPR-MDFIPGIDGPSEGVPR 764
>TC204787 similar to UP|Q84WK9 (Q84WK9) At4g33985, partial (61%)
Length = 977
Score = 29.6 bits (65), Expect = 6.1
Identities = 16/46 (34%), Positives = 25/46 (53%)
Frame = -2
Query: 316 VYIGDSVSYESTSSGLKSPLAPQPSSFCSGIDTPSEGLVRWKLRLE 361
V IGD+++ ++ L PLAP+P G+ P L W+ RL+
Sbjct: 307 VLIGDALA----AAVLAVPLAPEPRLIAEGVRRPRSLLEEWRGRLQ 182
>TC208310 weakly similar to UP|Q9MAL3 (Q9MAL3) F27F5.3, partial (52%)
Length = 847
Score = 29.3 bits (64), Expect = 8.0
Identities = 13/32 (40%), Positives = 20/32 (61%)
Frame = +3
Query: 326 STSSGLKSPLAPQPSSFCSGIDTPSEGLVRWK 357
STSS SP A + F +G D P EG+++++
Sbjct: 129 STSSSSPSPFATDQTVFVTGEDVPLEGVIQFE 224
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,020,490
Number of Sequences: 63676
Number of extensions: 496104
Number of successful extensions: 2647
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 2634
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2646
length of query: 889
length of database: 12,639,632
effective HSP length: 106
effective length of query: 783
effective length of database: 5,889,976
effective search space: 4611851208
effective search space used: 4611851208
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0252b.3


