

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252a.4
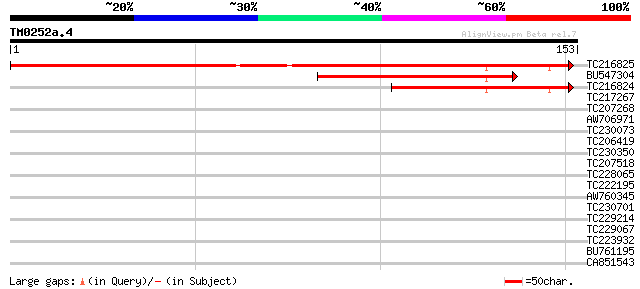
(153 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216825 similar to GB|AAB68983.1|500656|YSCH9666 Yhr143wp {Sacc... 248 1e-66
BU547304 92 1e-19
TC216824 77 2e-15
TC217267 similar to UP|Q8VZA5 (Q8VZA5) 3-deoxy-D-manno-octuloson... 32 0.16
TC207268 similar to UP|Q8GTM1 (Q8GTM1) WD40, partial (27%) 30 0.35
AW706971 similar to GP|13605607|gb| T3B23.2/T3B23.2 {Arabidopsis... 30 0.45
TC230073 similar to UP|Q9ASW6 (Q9ASW6) T3B23.2/T3B23.2, partial ... 30 0.45
TC206419 29 0.77
TC230350 weakly similar to UP|Q9C9F2 (Q9C9F2) MtN21-like protein... 28 1.7
TC207518 similar to UP|Q84M90 (Q84M90) At3g15180, partial (53%) 27 2.9
TC228065 similar to GB|AAO63313.1|28950779|BT005249 At4g16060 {A... 27 2.9
TC222195 27 3.8
AW760345 27 3.8
TC230701 weakly similar to UP|Q8W4C5 (Q8W4C5) Nucellin-like prot... 27 3.8
TC229214 similar to UP|Q42529 (Q42529) Tryptophan synthase alpha... 27 3.8
TC229067 27 5.0
TC223932 27 5.0
BU761195 27 5.0
CA851543 26 6.5
>TC216825 similar to GB|AAB68983.1|500656|YSCH9666 Yhr143wp {Saccharomyces
cerevisiae;} , partial (6%)
Length = 1557
Score = 248 bits (632), Expect = 1e-66
Identities = 127/160 (79%), Positives = 143/160 (89%), Gaps = 8/160 (5%)
Frame = +1
Query: 1 AIMYVLCFRMRSLVDIPRLKLQLLNMPMEPIWKHKLSPLKVCLPTVVEEFLRQAKAAQLF 60
AIMY+LCFRMRSL+DIPRLKLQLLNMPME I KHKLSPLKVCLPTVV EFLRQAKAAQLF
Sbjct: 556 AIMYILCFRMRSLMDIPRLKLQLLNMPMEAILKHKLSPLKVCLPTVVVEFLRQAKAAQLF 735
Query: 61 MSSSETFVFNDLLESDDLSRSFGGIDRLDMFFPFDPCLSKKSESYIRPHFVRWSRVRTTY 120
M +SE+FVF+D+LES DLS++FGG+DRLDMFFPFDPCL KKSESYIRPHFVRWS+VRTTY
Sbjct: 736 M-ASESFVFDDMLES-DLSKAFGGMDRLDMFFPFDPCLLKKSESYIRPHFVRWSKVRTTY 909
Query: 121 DSDDDED-----GSEMSHDDFVDRNAKEL---DMMVSVQG 152
D D+D D GSE+S DDF+DRNAK++ DMMVSV+G
Sbjct: 910 DDDNDNDEVSDSGSELSDDDFIDRNAKDMIDDDMMVSVEG 1029
>BU547304
Length = 526
Score = 91.7 bits (226), Expect = 1e-19
Identities = 43/58 (74%), Positives = 48/58 (82%), Gaps = 4/58 (6%)
Frame = -3
Query: 84 GIDRLDMFFPFDPCLSKKSESYIRPHFVRWSRVRTTYDSDDDED----GSEMSHDDFV 137
GIDRLDMFFPFDPCL KKSESYIRPHFVRWS+VRTTYD DD+++ GSE+S V
Sbjct: 524 GIDRLDMFFPFDPCLLKKSESYIRPHFVRWSKVRTTYDDDDNDELSDSGSEVSDHPLV 351
>TC216824
Length = 703
Score = 77.4 bits (189), Expect = 2e-15
Identities = 37/56 (66%), Positives = 46/56 (82%), Gaps = 7/56 (12%)
Frame = -1
Query: 104 SYIRPHFVRWSRVRTTYDSDDDED----GSEMSHDDFVDRNAKEL---DMMVSVQG 152
SYIRPHFVRWS+VRTTYD DD+++ GSE+S DDF+DRN K++ DMMVSV+G
Sbjct: 679 SYIRPHFVRWSKVRTTYDDDDNDELSDSGSEVSDDDFIDRNTKDMIDDDMMVSVEG 512
>TC217267 similar to UP|Q8VZA5 (Q8VZA5) 3-deoxy-D-manno-octulosonic acid
transferase-like protein, partial (24%)
Length = 1046
Score = 31.6 bits (70), Expect = 0.16
Identities = 14/36 (38%), Positives = 21/36 (57%)
Frame = +1
Query: 5 VLCFRMRSLVDIPRLKLQLLNMPMEPIWKHKLSPLK 40
+LC +R L + KL L + PM+ WKH + PL+
Sbjct: 490 ILCRFIRFLENWSSKKLSLSSSPMQHFWKHAVEPLR 597
>TC207268 similar to UP|Q8GTM1 (Q8GTM1) WD40, partial (27%)
Length = 790
Score = 30.4 bits (67), Expect = 0.35
Identities = 13/27 (48%), Positives = 21/27 (77%)
Frame = +2
Query: 18 RLKLQLLNMPMEPIWKHKLSPLKVCLP 44
R++ + LN+ ++P K+KL PLK+CLP
Sbjct: 314 RMRKRRLNLKLKP--KNKLMPLKICLP 388
>AW706971 similar to GP|13605607|gb| T3B23.2/T3B23.2 {Arabidopsis thaliana},
partial (31%)
Length = 422
Score = 30.0 bits (66), Expect = 0.45
Identities = 15/29 (51%), Positives = 18/29 (61%)
Frame = +1
Query: 101 KSESYIRPHFVRWSRVRTTYDSDDDEDGS 129
K SYI PHFV+ + D DDD+DGS
Sbjct: 277 KVNSYISPHFVK-----SNGDLDDDDDGS 348
>TC230073 similar to UP|Q9ASW6 (Q9ASW6) T3B23.2/T3B23.2, partial (93%)
Length = 863
Score = 30.0 bits (66), Expect = 0.45
Identities = 15/29 (51%), Positives = 18/29 (61%)
Frame = +3
Query: 101 KSESYIRPHFVRWSRVRTTYDSDDDEDGS 129
K SYI PHFV+ + D DDD+DGS
Sbjct: 585 KVNSYISPHFVK-----SNGDLDDDDDGS 656
>TC206419
Length = 1450
Score = 29.3 bits (64), Expect = 0.77
Identities = 15/56 (26%), Positives = 25/56 (43%)
Frame = +1
Query: 90 MFFPFDPCLSKKSESYIRPHFVRWSRVRTTYDSDDDEDGSEMSHDDFVDRNAKELD 145
+ P DP L + + +R +R ++ +DD+D HDDF + A D
Sbjct: 535 LHLPHDPALQRSLQLALRNRDDHDDDMRRSHHDEDDQDVEIDDHDDFEENCASHGD 702
>TC230350 weakly similar to UP|Q9C9F2 (Q9C9F2) MtN21-like protein;
91922-89607, partial (29%)
Length = 1057
Score = 28.1 bits (61), Expect = 1.7
Identities = 9/24 (37%), Positives = 16/24 (66%)
Frame = -2
Query: 91 FFPFDPCLSKKSESYIRPHFVRWS 114
F+P +PC + + ++PHF+R S
Sbjct: 501 FYPTEPCTAHTQSALLQPHFLRIS 430
>TC207518 similar to UP|Q84M90 (Q84M90) At3g15180, partial (53%)
Length = 1723
Score = 27.3 bits (59), Expect = 2.9
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 1/40 (2%)
Frame = -2
Query: 4 YVLCFRMRSLVDIPRLKLQL-LNMPMEPIWKHKLSPLKVC 42
Y L ++ L I R +L NMP IW H S LK+C
Sbjct: 1599 YCLTTQV*KLKSISRYHRRLSFNMPFNKIWTHSNSFLKLC 1480
>TC228065 similar to GB|AAO63313.1|28950779|BT005249 At4g16060 {Arabidopsis
thaliana;} , partial (91%)
Length = 1009
Score = 27.3 bits (59), Expect = 2.9
Identities = 14/38 (36%), Positives = 19/38 (49%)
Frame = -3
Query: 5 VLCFRMRSLVDIPRLKLQLLNMPMEPIWKHKLSPLKVC 42
+ CFR S K LN+P++ IW + S LK C
Sbjct: 548 IKCFRKNS-------KKPKLNLPLDKIWNLRFSNLKDC 456
>TC222195
Length = 844
Score = 26.9 bits (58), Expect = 3.8
Identities = 13/34 (38%), Positives = 20/34 (58%)
Frame = -3
Query: 116 VRTTYDSDDDEDGSEMSHDDFVDRNAKELDMMVS 149
V T D+DDD+D S S DD D + ++ ++ S
Sbjct: 275 VHDTEDADDDDDNSGHSPDDVDDDYSADVGLLGS 174
>AW760345
Length = 428
Score = 26.9 bits (58), Expect = 3.8
Identities = 14/29 (48%), Positives = 18/29 (61%)
Frame = +2
Query: 90 MFFPFDPCLSKKSESYIRPHFVRWSRVRT 118
+FFP PCLS S S++ P F SR R+
Sbjct: 5 LFFPICPCLSSLSRSHL-PSFQLRSRSRS 88
>TC230701 weakly similar to UP|Q8W4C5 (Q8W4C5) Nucellin-like protein, partial
(45%)
Length = 1119
Score = 26.9 bits (58), Expect = 3.8
Identities = 10/22 (45%), Positives = 15/22 (67%)
Frame = +2
Query: 31 IWKHKLSPLKVCLPTVVEEFLR 52
+W+ SP+ CL T++ EFLR
Sbjct: 356 LWRSTHSPIWSCLDTLIAEFLR 421
>TC229214 similar to UP|Q42529 (Q42529) Tryptophan synthase alpha chain ,
partial (84%)
Length = 1014
Score = 26.9 bits (58), Expect = 3.8
Identities = 11/24 (45%), Positives = 16/24 (65%)
Frame = -3
Query: 19 LKLQLLNMPMEPIWKHKLSPLKVC 42
LKL +L++P+E IW H+ L C
Sbjct: 970 LKLPVLSVPLEEIWPHQAPSL*HC 899
>TC229067
Length = 1161
Score = 26.6 bits (57), Expect = 5.0
Identities = 12/31 (38%), Positives = 18/31 (57%)
Frame = +1
Query: 13 LVDIPRLKLQLLNMPMEPIWKHKLSPLKVCL 43
LV ++K+ LLNM +P+ KH +CL
Sbjct: 508 LVTCHQMKILLLNMSFQPLEKHWNQDSPICL 600
>TC223932
Length = 530
Score = 26.6 bits (57), Expect = 5.0
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Frame = -2
Query: 4 YVLCFR-MRSLVDIPRLKLQLLNMPMEPIWKHKLSPLKVCLPTVVEEFLRQ 53
+ LCF M +LV I R ++ + PI+ H +K C+ VEE +R+
Sbjct: 424 FYLCFSLMYTLVIIVRSLIKTF*ETLFPIYYHAKIMMKFCIVIPVEERMRR 272
>BU761195
Length = 472
Score = 26.6 bits (57), Expect = 5.0
Identities = 15/56 (26%), Positives = 27/56 (47%)
Frame = -3
Query: 4 YVLCFRMRSLVDIPRLKLQLLNMPMEPIWKHKLSPLKVCLPTVVEEFLRQAKAAQL 59
+ LC + V +P++ + P+ KH++ P +PT E+ AK+ QL
Sbjct: 296 FFLCVNVSL*VLLPKVA------KLAPLCKHQMEPTNQPIPTSTEQKPENAKSPQL 147
>CA851543
Length = 633
Score = 26.2 bits (56), Expect = 6.5
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Frame = +2
Query: 98 LSKKSESYIRPHFVRW--SRVRTTYDSDDDEDGSEMSHDDFVDRNAKELD 145
+S + S +P +W RV + +DD DG+E+S D V + +LD
Sbjct: 473 ISSRRRSQPKPDHSKWVICRVYERDNDEDDGDGTELSCLDEVFLSLDDLD 622
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.139 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,592,831
Number of Sequences: 63676
Number of extensions: 82071
Number of successful extensions: 704
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 694
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 698
length of query: 153
length of database: 12,639,632
effective HSP length: 89
effective length of query: 64
effective length of database: 6,972,468
effective search space: 446237952
effective search space used: 446237952
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0252a.4


