

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252a.3
(282 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
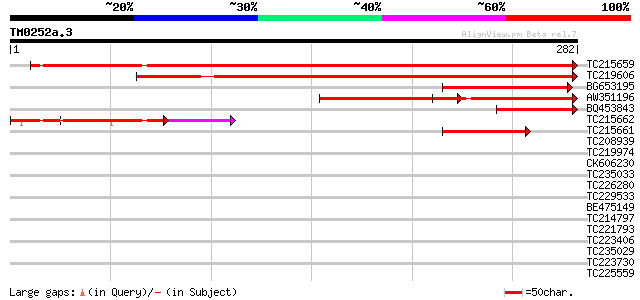
TC215659 homologue to UP|Q94FX1 (Q94FX1) Heme oxygenase 1 (Fragm... 420 e-118
TC219606 similar to UP|O48722 (O48722) Heme oxygenase 2 precurso... 241 3e-64
BG653195 GP|14485569|gb heme oxygenase 3 {Glycine max}, partial ... 117 4e-27
AW351196 similar to GP|14485567|gb heme oxygenase 1 {Glycine max... 83 2e-16
BQ453843 GP|14485567|gb heme oxygenase 1 {Glycine max}, partial ... 75 4e-14
TC215662 homologue to UP|Q94FX0 (Q94FX0) Heme oxygenase 3 (Fragm... 67 1e-11
TC215661 similar to UP|Q94FX0 (Q94FX0) Heme oxygenase 3 (Fragmen... 60 8e-10
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 30 0.94
TC219974 weakly similar to PIR|T47938|T47938 RING finger protein... 30 1.2
CK606230 30 1.6
TC235033 25 3.5
TC226280 similar to UP|EF1G_PRUAV (Q9FUM1) Elongation factor 1-g... 28 3.6
TC229533 similar to UP|Q6NLA5 (Q6NLA5) At5g03080, partial (55%) 28 4.7
BE475149 28 4.7
TC214797 homologue to UP|R15D_ARATH (Q9FY64) 40S ribosomal prote... 28 4.7
TC221793 similar to UP|CENB_HUMAN (P07199) Major centromere auto... 28 6.1
TC223406 weakly similar to UP|Q9FG99 (Q9FG99) Similarity to GTP-... 28 6.1
TC235029 similar to GB|CAA54234.1|472869|ATARPMR ARP protein {Ar... 28 6.1
TC223730 similar to UP|Q7SIC9 (Q7SIC9) Transferase, partial (29%) 23 6.8
TC225559 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 27 8.0
>TC215659 homologue to UP|Q94FX1 (Q94FX1) Heme oxygenase 1 (Fragment),
complete
Length = 1365
Score = 420 bits (1079), Expect = e-118
Identities = 209/272 (76%), Positives = 234/272 (85%)
Frame = +1
Query: 11 QSLYIIKPRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETARKRYPGE 70
QSLYI KPRL+P P P QFRS++ + + ++ VIVSA AET +K+ GE
Sbjct: 427 QSLYI-KPRLAPRPLPPPQFRSLFLTSPAGARTATVMPRRAAVIVSAATAETPKKK--GE 597
Query: 71 SKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDTL 130
SKGFVEEMRFVAM+LHT+DQA+EGEKEV +PEEKAVTKW P+V+GYLKFLVDSK+VYDTL
Sbjct: 598 SKGFVEEMRFVAMRLHTRDQAREGEKEVKQPEEKAVTKWNPSVEGYLKFLVDSKLVYDTL 777
Query: 131 EKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQN 190
EKIV +A HP YAEFRNTGLERSASL +DL+WFKEQGYTIP+PSSPGL YAQYL++LS
Sbjct: 778 EKIVHEAPHPFYAEFRNTGLERSASLVEDLDWFKEQGYTIPEPSSPGLTYAQYLKELSVK 957
Query: 191 DPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKV 250
DPQAFICHFYNIYFAHSAGGRMIGKK+A +LLNNK LEFYKWDGDL QLLQNVRDKLNKV
Sbjct: 958 DPQAFICHFYNIYFAHSAGGRMIGKKVAEKLLNNKALEFYKWDGDLPQLLQNVRDKLNKV 1137
Query: 251 AEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
AE WTREEK+HCLEETEKSFK SGEILRLILS
Sbjct: 1138AEPWTREEKDHCLEETEKSFKLSGEILRLILS 1233
>TC219606 similar to UP|O48722 (O48722) Heme oxygenase 2 precursor (HO2),
partial (66%)
Length = 948
Score = 241 bits (615), Expect = 3e-64
Identities = 112/219 (51%), Positives = 158/219 (72%)
Frame = +2
Query: 64 RKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDS 123
RK YPGE+ G EEMRFVAM+L T D + E + W+ +++G+L +LVDS
Sbjct: 266 RKLYPGETTGITEEMRFVAMRLRTNDTVSQQE------HQSHSDAWQASMEGFLSYLVDS 427
Query: 124 KVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQY 183
+++ TL++IV ++ + SYA R TGLERS L KDL+W +E+G IP P SPGL YA+Y
Sbjct: 428 HLIFATLQRIVDESDNVSYAYMRKTGLERSEGLLKDLKWLEEEGNMIPTPGSPGLTYAKY 607
Query: 184 LRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNV 243
L +L++ F+ HFYNIYF+H A G++IGKK++ ELL K LEFYKW+GD+ LL++V
Sbjct: 608 LEELAEISAPLFLSHFYNIYFSHIAAGQVIGKKVSEELLEGKELEFYKWEGDVPDLLKDV 787
Query: 244 RDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
RDKLN ++E W+R+EKN CL+ET K+F++ G+I+RLI+S
Sbjct: 788 RDKLNMLSEHWSRDEKNRCLKETTKAFRYMGQIVRLIVS 904
>BG653195 GP|14485569|gb heme oxygenase 3 {Glycine max}, partial (25%)
Length = 382
Score = 117 bits (294), Expect = 4e-27
Identities = 56/65 (86%), Positives = 60/65 (92%)
Frame = +3
Query: 216 KIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGE 275
++A +LLNNK LEFYKWDGDL QLLQNVRDKLNKVAE WTREEK+HCLEETEKSFK SGE
Sbjct: 186 QVAEKLLNNKALEFYKWDGDLPQLLQNVRDKLNKVAEPWTREEKDHCLEETEKSFKLSGE 365
Query: 276 ILRLI 280
ILRLI
Sbjct: 366 ILRLI 380
>AW351196 similar to GP|14485567|gb heme oxygenase 1 {Glycine max}, partial
(49%)
Length = 471
Score = 82.8 bits (203), Expect = 2e-16
Identities = 41/71 (57%), Positives = 50/71 (69%)
Frame = -1
Query: 155 SLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIG 214
S+AKD EWFKEQG I PG+ A+YL++ S DPQAFI HF +IY+ HS GGR IG
Sbjct: 468 SVAKDWEWFKEQGXXIRGHYYPGIT*ARYLKE*SAKDPQAFISHFSHIYWDHSGGGRTIG 289
Query: 215 KKIAGELLNNK 225
KK+A +LL K
Sbjct: 288 KKVAEKLLXXK 256
Score = 80.1 bits (196), Expect = 1e-15
Identities = 40/72 (55%), Positives = 52/72 (71%)
Frame = -2
Query: 211 RMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSF 270
R+ + + L +G+ +KWD DL +LLQNVR KLNKV E W+REEK+HC E+TE+SF
Sbjct: 296 RLEKRSLKSYLXXRRGV--HKWDDDLPRLLQNVRXKLNKVVEPWSREEKDHCPEQTEQSF 123
Query: 271 KWSGEILRLILS 282
K GEILR I+S
Sbjct: 122 KLCGEILREIVS 87
>BQ453843 GP|14485567|gb heme oxygenase 1 {Glycine max}, partial (15%)
Length = 413
Score = 74.7 bits (182), Expect = 4e-14
Identities = 35/40 (87%), Positives = 38/40 (94%)
Frame = +1
Query: 243 VRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
+RDKLNKVAE W+REEK+HCLEETEKSFK SGEILRLILS
Sbjct: 250 IRDKLNKVAEPWSREEKDHCLEETEKSFKLSGEILRLILS 369
>TC215662 homologue to UP|Q94FX0 (Q94FX0) Heme oxygenase 3 (Fragment),
partial (31%)
Length = 392
Score = 66.6 bits (161), Expect = 1e-11
Identities = 38/87 (43%), Positives = 50/87 (56%)
Frame = +2
Query: 26 PHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKL 85
PHR++R + F L+ R +S R+R KG + RF AM+L
Sbjct: 161 PHRRWR-VVFVLLSCLEGLR---------LSCRRRRRRRRRRRASRKGSWKR*RFAAMRL 310
Query: 86 HTKDQAKEGEKEVTEPEEKAVTKWEPT 112
HT+DQA+EGEKEV +PEEKAVTKW P+
Sbjct: 311 HTRDQAREGEKEVKQPEEKAVTKWNPS 391
Score = 63.9 bits (154), Expect = 8e-11
Identities = 44/83 (53%), Positives = 56/83 (67%), Gaps = 4/83 (4%)
Frame = +1
Query: 1 MASA--TVVSQIQSLYIIKPRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQM--KSVVIVS 56
MAS+ T +SQ QSLYI KPRL+P PP H QFRS++ + + R M ++ VIVS
Sbjct: 55 MASSLTTTLSQSQSLYI-KPRLAPRPP-HPQFRSLFLTSPAVAGGVRAAVMPRRTAVIVS 228
Query: 57 ATAAETARKRYPGESKGFVEEMR 79
A AET +K+ GESKGFVEEM+
Sbjct: 229 AATAETPKKK--GESKGFVEEMK 291
>TC215661 similar to UP|Q94FX0 (Q94FX0) Heme oxygenase 3 (Fragment), partial
(16%)
Length = 704
Score = 60.5 bits (145), Expect = 8e-10
Identities = 28/44 (63%), Positives = 35/44 (78%)
Frame = +3
Query: 216 KIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEK 259
++A +LLNN L FYKW GDL LLQNV++KLNKVAE TR++K
Sbjct: 546 QVAEKLLNNXALXFYKWXGDLPPLLQNVKEKLNKVAEPGTRKKK 677
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 30.4 bits (67), Expect = 0.94
Identities = 23/80 (28%), Positives = 37/80 (45%), Gaps = 12/80 (15%)
Frame = +3
Query: 70 ESKGFVEEMRFVAMKLHTKD-------QAKEGEKEVTEPEE---KAVTKWEPTVDGYLKF 119
E K +M+ V K+ KD + K+GE +V PEE +TK + T + +L
Sbjct: 393 EDKEVQRDMKLVPYKIVNKDGKPYIQEKIKDGETKVFSPEEISAMILTKMKETAEAFLGK 572
Query: 120 LVDSKVVY--DTLEKIVQDA 137
++ V Y D + +DA
Sbjct: 573 KINDAVAYFNDAQRQATKDA 632
>TC219974 weakly similar to PIR|T47938|T47938 RING finger protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(64%)
Length = 690
Score = 30.0 bits (66), Expect = 1.2
Identities = 19/52 (36%), Positives = 26/52 (49%), Gaps = 1/52 (1%)
Frame = +3
Query: 18 PRLSP-PPPPHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETARKRYP 68
PRL P PPPPH + +HRFR++ SAT A++ R +P
Sbjct: 201 PRLRPLPPPPHLR-----------PPRHRFRRL*PPSAASATRADSLRPPHP 323
>CK606230
Length = 615
Score = 29.6 bits (65), Expect = 1.6
Identities = 22/75 (29%), Positives = 35/75 (46%), Gaps = 10/75 (13%)
Frame = +1
Query: 3 SATVVSQIQSLYIIKPRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQMKSV---------- 52
SAT+ + S I++ RL P P ++ F R ++HRFRQ S
Sbjct: 157 SATL*TGSPSPIILRLRLLHPRPSRHHSLALQFQGRRRRRRHRFRQNPSFCHSSRRDSTS 336
Query: 53 VIVSATAAETARKRY 67
++ S+ A++ AR Y
Sbjct: 337 LLFSSQASQGARNNY 381
>TC235033
Length = 404
Score = 25.0 bits (53), Expect(2) = 3.5
Identities = 9/10 (90%), Positives = 9/10 (90%)
Frame = +2
Query: 17 KPRLSPPPPP 26
KP LSPPPPP
Sbjct: 209 KPPLSPPPPP 238
Score = 21.9 bits (45), Expect(2) = 3.5
Identities = 10/25 (40%), Positives = 14/25 (56%)
Frame = +3
Query: 23 PPPPHRQFRSIYFPTTRLLQQHRFR 47
P PPH R P +R L++H+ R
Sbjct: 321 PQPPHPPLRPPLLPPSR-LRRHQLR 392
>TC226280 similar to UP|EF1G_PRUAV (Q9FUM1) Elongation factor 1-gamma
(EF-1-gamma) (eEF-1B gamma), complete
Length = 1703
Score = 28.5 bits (62), Expect = 3.6
Identities = 29/100 (29%), Positives = 43/100 (43%)
Frame = +3
Query: 34 YFPTTRLLQQHRFRQMKSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKE 93
YF T L+ Q FR++ V + + P +SK + K K AK
Sbjct: 732 YFWT--LVNQPNFRKILGQVKQTEAVPPIPSAKKPSQSKESKPKP-----KDEPKKVAKP 890
Query: 94 GEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDTLEKI 133
KEV EPEE+A +P L L SK++ D +++
Sbjct: 891 EPKEVAEPEEEAP---KPKPKNPLDLLPPSKMILDEWKRL 1001
>TC229533 similar to UP|Q6NLA5 (Q6NLA5) At5g03080, partial (55%)
Length = 888
Score = 28.1 bits (61), Expect = 4.7
Identities = 13/33 (39%), Positives = 19/33 (57%)
Frame = +2
Query: 9 QIQSLYIIKPRLSPPPPPHRQFRSIYFPTTRLL 41
Q + L++ + R PPPPPH R+ + RLL
Sbjct: 137 QRRPLFLARARQPPPPPPHVVPRAAHHVFPRLL 235
>BE475149
Length = 359
Score = 28.1 bits (61), Expect = 4.7
Identities = 18/50 (36%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Frame = -3
Query: 56 SATAAETARKRYPGESK-GFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEK 104
+ATAA T + E K GF+++++ K+H + A EGEK+ E ++K
Sbjct: 282 AATAAPTIHGEHKPEHKEGFLDKVKD---KIHGEGGAAEGEKKKKEKKKK 142
>TC214797 homologue to UP|R15D_ARATH (Q9FY64) 40S ribosomal protein S15-4,
partial (95%)
Length = 798
Score = 28.1 bits (61), Expect = 4.7
Identities = 11/32 (34%), Positives = 17/32 (52%)
Frame = -3
Query: 22 PPPPPHRQFRSIYFPTTRLLQQHRFRQMKSVV 53
PPPPPH+ R + P R+L + + + V
Sbjct: 109 PPPPPHQPQRLPFSPAARVLASNNNNKQEKAV 14
>TC221793 similar to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (3%)
Length = 801
Score = 27.7 bits (60), Expect = 6.1
Identities = 18/67 (26%), Positives = 32/67 (46%)
Frame = -3
Query: 50 KSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKW 109
+SVV V+ A G++ G + + K +++ EGE+E E EE +W
Sbjct: 382 RSVVAVAGEAVIPENFNRSGDAVG-AGRVPEGSEKSEEENEEGEGEEEA*EDEEGGSARW 206
Query: 110 EPTVDGY 116
+V+G+
Sbjct: 205 RASVEGF 185
>TC223406 weakly similar to UP|Q9FG99 (Q9FG99) Similarity to GTP-binding
regulatory protein and WD-repeat protein, partial (14%)
Length = 422
Score = 27.7 bits (60), Expect = 6.1
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 5/26 (19%)
Frame = +1
Query: 13 LYIIKPRLSP-----PPPPHRQFRSI 33
L+ +PRLSP PPPPH +F +
Sbjct: 121 LFTTQPRLSPLPQPPPPPPHPEFHRL 198
>TC235029 similar to GB|CAA54234.1|472869|ATARPMR ARP protein {Arabidopsis
thaliana;} , partial (28%)
Length = 625
Score = 27.7 bits (60), Expect = 6.1
Identities = 17/54 (31%), Positives = 26/54 (47%)
Frame = +3
Query: 106 VTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKD 159
VT+W+P++ YLK L SK V T + + H + G +RSA +
Sbjct: 51 VTEWDPSLSNYLKELEKSKPVILTGD---LNCAHEEIDIYNPAGNKRSAGFTDE 203
>TC223730 similar to UP|Q7SIC9 (Q7SIC9) Transferase, partial (29%)
Length = 865
Score = 23.1 bits (48), Expect(2) = 6.8
Identities = 8/9 (88%), Positives = 8/9 (88%)
Frame = +2
Query: 18 PRLSPPPPP 26
P LSPPPPP
Sbjct: 47 PPLSPPPPP 73
Score = 22.7 bits (47), Expect(2) = 6.8
Identities = 11/24 (45%), Positives = 11/24 (45%)
Frame = +3
Query: 22 PPPPPHRQFRSIYFPTTRLLQQHR 45
PPPPPH P RL Q R
Sbjct: 123 PPPPPHH-----LHPRRRLRQNRR 179
>TC225559 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (47%)
Length = 870
Score = 27.3 bits (59), Expect = 8.0
Identities = 9/18 (50%), Positives = 14/18 (77%)
Frame = +1
Query: 9 QIQSLYIIKPRLSPPPPP 26
Q+Q L++++ L PPPPP
Sbjct: 256 QLQPLHLLRRHLPPPPPP 309
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,498,438
Number of Sequences: 63676
Number of extensions: 171972
Number of successful extensions: 2281
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 2065
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2263
length of query: 282
length of database: 12,639,632
effective HSP length: 96
effective length of query: 186
effective length of database: 6,526,736
effective search space: 1213972896
effective search space used: 1213972896
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0252a.3


