

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0250.10
(1065 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
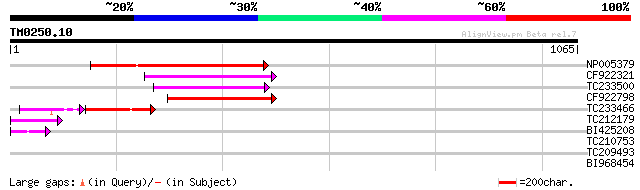
Score E
Sequences producing significant alignments: (bits) Value
NP005379 ORF (334 AA) 336 3e-92
CF922321 229 6e-60
TC233500 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable... 204 2e-52
CF922798 192 5e-49
TC233466 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable... 91 7e-31
TC212179 72 2e-12
BI425208 49 9e-06
TC210753 similar to UP|Q9SAF1 (Q9SAF1) F3F19.20 protein (Actin-r... 32 1.9
TC209493 similar to UP|Q8VXU5 (Q8VXU5) AT3g53630/F4P12_330, part... 30 5.7
BI968454 similar to GP|13171103|gb|A rRNA intron-encoded homing ... 30 7.4
>NP005379 ORF (334 AA)
Length = 1002
Score = 336 bits (862), Expect = 3e-92
Identities = 159/335 (47%), Positives = 220/335 (65%), Gaps = 1/335 (0%)
Frame = +1
Query: 153 HGWSNVSFTALLELLKEAMP-DLNIPESFNKTKGMIRDLGLDYKRIHACPNDCMLYWKEY 211
+GWS+ SFT+LL+++ + +P D +P+S+ + K ++ +G++Y++IHACPNDC+LY E+
Sbjct: 1 YGWSDKSFTSLLQIVHDLLPQDNTLPKSYYQAKKILCPMGMEYQKIHACPNDCILYRHEF 180
Query: 212 ENDNSCRTCKASRWKEFPQVAGEFEKSESEYKVPAKVLRHFPLIPRLQRLFMCSRTSDSM 271
E + C C ASR+K +++ S+ PAKVL + P+IPR +RLF + +
Sbjct: 181 EEMSKCPRCGASRYKVKDDEDCSSDEN-SKKGPPAKVLWYLPIIPRFKRLFANEDDAKDL 357
Query: 272 RWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHST 331
WH R DG +RHP D W+ D L+ +F + RN+RLGLASDG NP+ +S HS+
Sbjct: 358 TWHANGRKSDGMVRHPADCSQWKKIDSLYPNFGKEARNLRLGLASDGMNPYGNLSTQHSS 537
Query: 332 WPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYD 391
WPVLLV+YN+PPWLCMK +YMM+S++I G + PG +IDVYL PLIE+L++LW+ GV +D
Sbjct: 538 WPVLLVIYNFPPWLCMKRKYMMLSMMISGPRQPGNDIDVYLSPLIEDLRKLWDEGVLVFD 717
Query: 392 ASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMDH 451
RKETFQM A L TIND+PAY LSG+S KG LAC C +TS LKH K Y H
Sbjct: 718 GFRKETFQMRAMLFCTINDFPAYGNLSGYSVKGHLACPICEEDTSYIQLKHGRKTVYTRH 897
Query: 452 RVFLPADHTWRTTENLFNGKMEFRTASPLLKGTEI 486
RVFL A H +R + FNG E L G ++
Sbjct: 898 RVFLKAHHPYRRLKKAFNGSQEHEIRRTPLTGEQV 1002
>CF922321
Length = 742
Score = 229 bits (583), Expect = 6e-60
Identities = 111/247 (44%), Positives = 148/247 (58%)
Frame = -1
Query: 254 LIPRLQRLFMCSRTSDSMRWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLG 313
+IPRL+RLF + + WH R+ DG +R W+ D L+LD + N+RLG
Sbjct: 742 IIPRLKRLFANGDDAKDLTWHANGRNCDGMIRRLAVSSQWKKIDRLYLDLGKEASNLRLG 563
Query: 314 LASDGFNPFRTMSISHSTWPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQ 373
LASDG NP+ + S HS+WPVLLV+YN PPWLCMK +YMM+S++I + PG +IDVYL
Sbjct: 562 LASDGMNPYGSSST*HSSWPVLLVIYNLPPWLCMKRKYMMLSMMISNPRQPGNDIDVYLS 383
Query: 374 PLIEELKELWEVGVETYDASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLACACCNY 433
LIE+L +LW+ G ++ ETF M A L TIND+PAY LSG++ KG +C C
Sbjct: 382 SLIEDLTKLWDEGFLAFNGFWNETFPMRAMLFCTINDFPAYGNLSGYNVKGHHSCPICEE 203
Query: 434 NTSSTYLKHSHKMCYMDHRVFLPADHTWRTTENLFNGKMEFRTASPLLKGTEILEILKNF 493
NTS LKH K Y HR FL +H +R + FNG L E+ + +++
Sbjct: 202 NTSYIQLKHGRKTVYSRHRRFLTPNHPYRRLKKAFNGSQGHDIVPIPLTAEEVYQRVQHL 23
Query: 494 NNVFGKT 500
NNV GKT
Sbjct: 22 NNVLGKT 2
>TC233500 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable element
(Fragment), partial (65%)
Length = 1033
Score = 204 bits (518), Expect = 2e-52
Identities = 98/218 (44%), Positives = 131/218 (59%)
Frame = +2
Query: 271 MRWHEEERSKDGKLRHPVDGQAWQDFDGLHLDFASDPRNVRLGLASDGFNPFRTMSISHS 330
+ WH R+ D LRH D W+ D L+ DF + +N+ LGLA+D NP+ +S H+
Sbjct: 8 LTWHANGRNCDRMLRHLADSLQWKKIDRLYPDFGKETKNLTLGLATDAMNPYGCLSTQHN 187
Query: 331 TWPVLLVVYNYPPWLCMKSEYMMMSLLIPGQQSPGKNIDVYLQPLIEELKELWEVGVETY 390
+WPVLLV+YN P WLCMK +YMM+S++I G + PG +I+VYL PLIE+L +L + GV +
Sbjct: 188 SWPVLLVIYNLPHWLCMKRKYMMLSMMITGPRQPGNDINVYLNPLIEDLTKL*DEGVLVF 367
Query: 391 DASRKETFQMHAALLWTINDYPAYAMLSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMD 450
D R ETF + A L TIND+P Y LSG+S KG AC C +TS KH K Y
Sbjct: 368 DGFRNETFNLCAMLFCTINDFPTYENLSGYSVKGHRACPICEEDTSYIQQKHGRKTVYTR 547
Query: 451 HRVFLPADHTWRTTENLFNGKMEFRTASPLLKGTEILE 488
HR FL H +R + FN E A LL G ++ +
Sbjct: 548 HRRFLKPHHPYRRLKKAFNES*EHENALVLLTGDQVFQ 661
>CF922798
Length = 617
Score = 192 bits (489), Expect = 5e-49
Identities = 94/204 (46%), Positives = 127/204 (62%)
Frame = -2
Query: 297 DGLHLDFASDPRNVRLGLASDGFNPFRTMSISHSTWPVLLVVYNYPPWLCMKSEYMMMSL 356
D L+LDF+ + N+RLGLA+DG NP+ ++S +S W VLLV+YN PWLCM +YMM+S+
Sbjct: 613 DHLYLDFSKEAINLRLGLATDGMNPYGSLSTQYSLWSVLLVIYNLSPWLCMNKKYMMLSM 434
Query: 357 LIPGQQSPGKNIDVYLQPLIEELKELWEVGVETYDASRKETFQMHAALLWTINDYPAYAM 416
+I + PG +IDVYL PLIE+L L + V +D + ETF + A L TIND+PAY
Sbjct: 433 MILSPRQPGNDIDVYLSPLIEDLTML*DEEVLVFDGFQNETFHLCAMLFCTINDFPAYGN 254
Query: 417 LSGWSTKGKLACACCNYNTSSTYLKHSHKMCYMDHRVFLPADHTWRTTENLFNGKMEFRT 476
LSG+S K AC C +TS LKH K Y H+ FL H +R + FNG E T
Sbjct: 253 LSGYSVKSHRACPICEEDTSYIQLKHGRKTIYTRHQRFLKPHHLYRRLKKEFNGSQEHET 74
Query: 477 ASPLLKGTEILEILKNFNNVFGKT 500
LL G + + +++ N +FGKT
Sbjct: 73 TPILLVGDHVYQQVQHLNTIFGKT 2
>TC233466 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable element
(Fragment), partial (37%)
Length = 790
Score = 90.5 bits (223), Expect(2) = 7e-31
Identities = 45/134 (33%), Positives = 82/134 (60%), Gaps = 2/134 (1%)
Frame = +2
Query: 143 IIRMYLLKCTHGWSNVSFTALLELLKEAMPDLN-IPESFNKTKGMIRDLGLDYKRIHACP 201
++ + +K +G S+ SF++LL+++ + +P+ N +P+S+ + K ++ + ++Y +IHACP
Sbjct: 386 VLSLVNMKARYGCSDKSFSSLLQVVHDVLPEENTLPKSYYQAKKILCSMDMEY*KIHACP 565
Query: 202 NDCMLYWKEYENDNSCRTCKASRWKEFPQVAGEFEKSESEYKVP-AKVLRHFPLIPRLQR 260
NDC+LY E+E + C C SR+K + E E+ K P AKVL + P++P+ +R
Sbjct: 566 NDCILYRHEFEEMSKCPRCGVSRYK--VKDDEECSSDENSKKGPLAKVLWYLPIVPKFKR 739
Query: 261 LFMCSRTSDSMRWH 274
LF + + WH
Sbjct: 740 LFANGDDAKDLTWH 781
Score = 63.2 bits (152), Expect(2) = 7e-31
Identities = 44/132 (33%), Positives = 57/132 (42%), Gaps = 9/132 (6%)
Frame = +3
Query: 18 GVESFLDFAYTKGRP-QGDEILCPCAKCGNCCWEKRNVIYNHLICTGFLEGYNVWIHHGE 76
GVE FL A + RP + + CPC C N +K N I +HL+C Y WI HGE
Sbjct: 12 GVEQFLQLASERSRPDENGKFFCPCINCLNGRRQKVNDIRDHLLCDEIKRNYTTWIWHGE 191
Query: 77 --------QMMEIDEEMEEDYHDDIDGLLYETFKYVAGAEPEGPNEDARKFYNLIDESQK 128
Q D EM + D I L E+F+ + P D + +S+
Sbjct: 192 MADMQSGPQSEPFDVEMRDRLEDMIRDLGQESFQ-----QAHAPMYDTLQI-----DSKT 341
Query: 129 ELYP*CKNFSTL 140
LYP C N TL
Sbjct: 342 PLYPGCNNLLTL 377
>TC212179
Length = 764
Score = 71.6 bits (174), Expect = 2e-12
Identities = 37/99 (37%), Positives = 48/99 (48%), Gaps = 1/99 (1%)
Frame = +1
Query: 1 MDKEWTKQPRFSSEYINGVESFLDFAYTKGRPQGD-EILCPCAKCGNCCWEKRNVIYNHL 59
MD+ W Q R S EY GVE FL FA +G+P D + CPC C N + + I HL
Sbjct: 31 MDRSWMNQSRISPEYEEGVEQFLQFASQRGQPDEDGKYYCPCINCLNGRRQILDDIREHL 210
Query: 60 ICTGFLEGYNVWIHHGEQMMEIDEEMEEDYHDDIDGLLY 98
+C G Y WI HGE + E + ++ LY
Sbjct: 211 LCDGIKRNYTTWIWHGEMTDMQSGQQSEPFDVEMGDHLY 327
>BI425208
Length = 413
Score = 49.3 bits (116), Expect = 9e-06
Identities = 23/76 (30%), Positives = 42/76 (55%), Gaps = 1/76 (1%)
Frame = -3
Query: 2 DKEWTK-QPRFSSEYINGVESFLDFAYTKGRPQGDEILCPCAKCGNCCWEKRNVIYNHLI 60
+K W K + S EY+ G+ FL+F+++ + + + I CPC +C ++ R+ + L+
Sbjct: 228 NKGWIKLNIQASIEYLKGINDFLEFSFSYSKNK-NTIRCPCCRCNAIYYKTRDEVIGDLL 52
Query: 61 CTGFLEGYNVWIHHGE 76
+ F + Y W HGE
Sbjct: 51 KSRFQQNYEHWESHGE 4
>TC210753 similar to UP|Q9SAF1 (Q9SAF1) F3F19.20 protein (Actin-related
protein 3) (At1g13180), partial (8%)
Length = 1210
Score = 31.6 bits (70), Expect = 1.9
Identities = 18/46 (39%), Positives = 22/46 (47%), Gaps = 2/46 (4%)
Frame = +3
Query: 55 IYNHLICTGFLEGYNVWIHHGEQMMEIDEEMEEDYHDDIDG--LLY 98
I +HLIC G + Y WI HGE E ++DI LLY
Sbjct: 300 IRSHLICDGIVPTYIKWI*HGESSNMATVSQAEPVNEDIGNRFLLY 437
>TC209493 similar to UP|Q8VXU5 (Q8VXU5) AT3g53630/F4P12_330, partial (46%)
Length = 1023
Score = 30.0 bits (66), Expect = 5.7
Identities = 27/88 (30%), Positives = 41/88 (45%), Gaps = 12/88 (13%)
Frame = +3
Query: 55 IYNHLICTGFLE----GYNVWIHHGEQMMEIDEEMEEDYHDD-IDGLLYETFK------- 102
+ +H IC GF + GY VW HG +M DEE Y+ + I G ++ FK
Sbjct: 333 VRDHHICVGFQDEEGSGYRVWEWHG-HIMAFDEEF--GYNPEYIYGNYFQWFKARPFSVA 503
Query: 103 YVAGAEPEGPNEDARKFYNLIDESQKEL 130
VA A + E+ + L ++ +EL
Sbjct: 504 AVADARADVEKEEEEEEEKLENQGLREL 587
>BI968454 similar to GP|13171103|gb|A rRNA intron-encoded homing endonuclease
{Oryza sativa}, partial (60%)
Length = 396
Score = 29.6 bits (65), Expect = 7.4
Identities = 17/55 (30%), Positives = 25/55 (44%), Gaps = 9/55 (16%)
Frame = +1
Query: 211 YENDNSCRTCKASRWKEFP---------QVAGEFEKSESEYKVPAKVLRHFPLIP 256
Y N N+ +T RW E+P + K+ +KVPA+ + PLIP
Sbjct: 40 YRNGNAQKTTSKERWNEWP*AFSHLRGLTTSPRNPKTLISHKVPAEP*KQHPLIP 204
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.348 0.153 0.544
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,521,758
Number of Sequences: 63676
Number of extensions: 905588
Number of successful extensions: 9098
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 6710
Number of HSP's successfully gapped in prelim test: 287
Number of HSP's that attempted gapping in prelim test: 2175
Number of HSP's gapped (non-prelim): 7315
length of query: 1065
length of database: 12,639,632
effective HSP length: 107
effective length of query: 958
effective length of database: 5,826,300
effective search space: 5581595400
effective search space used: 5581595400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0250.10


