

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0240.16
(1549 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
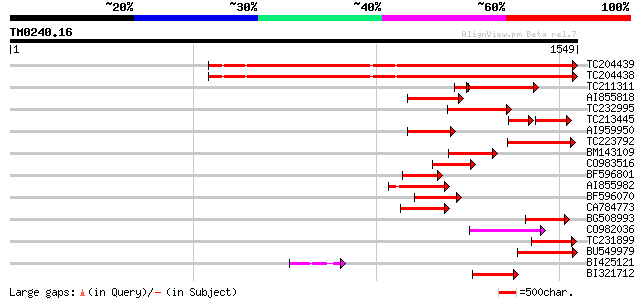
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 960 0.0
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 958 0.0
TC211311 weakly similar to UP|O24587 (O24587) Pol protein, parti... 207 9e-63
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 221 1e-57
TC232995 213 4e-55
TC213445 129 1e-48
AI959950 166 1e-40
TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polypro... 162 1e-39
BM143109 155 1e-37
CO983516 152 8e-37
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 149 7e-36
AI855982 142 1e-33
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 131 2e-30
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 123 5e-28
BG508993 120 4e-27
CO982036 120 4e-27
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 120 6e-27
BU549979 118 2e-26
BI425121 113 7e-25
BI321712 110 5e-24
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 960 bits (2481), Expect = 0.0
Identities = 487/1006 (48%), Positives = 666/1006 (65%)
Frame = +1
Query: 544 LLQTSYAAQLRHLSWYLDSGCSRHMTGTRSIFQKLTPLKSGGDVGFGGNQKGKIIGKGSI 603
++ TS A + WYLDSGCSRHMTG + + P S V FG KGKIIG G +
Sbjct: 1645 VVHTSLRASAKE-DWYLDSGCSRHMTGVKEFLLNIEPC-STSYVTFGDGSKGKIIGMGKL 1818
Query: 604 GDGKTPVINDVLLVEGLFHNLLSISQIADKGYDVIFNQTGCKAVSQTDGSVLFSGKRKND 663
P +N VLLV+GL NL+SISQ+ D+G++V F ++ C V+ VL G R D
Sbjct: 1819 VHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKD 1995
Query: 664 IYKIRSSELLSQKVKCLMSVNDEQWIWHRRLGHASLRKISQLSKLNLVRGLPHLKYSSEA 723
+ + + S CL S DE IWH+R GH LR + ++ VRG+P+LK
Sbjct: 1996 NCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGR 2175
Query: 724 LCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYELVIVDDYSRWT 783
+C CQ GK K + +T+R LELLH+DL GP++ ES+GGK+Y V+VDD+SR+T
Sbjct: 2176 ICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFT 2355
Query: 784 WVKFLRHKDETHTMFTNFITQVQKEFQSSVITVRSDHGGEFENKAFEDLFNSQGISHNFS 843
WV F+R K ET +F ++Q+E + +RSDHG EFEN F + S+GI+H FS
Sbjct: 2356 WVNFIREKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFS 2535
Query: 844 CPRTPQQNGVVERKNITLQEMARTMMQESSMAKHFWAEAINTACYIQNRISIRPILEKTP 903
TPQQNG+VERKN TLQE AR M+ + + WAEA+NTACYI NR+++R T
Sbjct: 2536 AAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTL 2715
Query: 904 YELCKGRQPDISYFHPFGSTCFMLNTKEQLGKFDSKALKCYFLGYSERSKGFRIYNIIHQ 963
YE+ KGR+P + +FH FGS C++L +EQ K D K+ FLGYS S+ +R++N +
Sbjct: 2716 YEIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTR 2895
Query: 964 TVEESIQIRFDDKLGSEKSKLFERFADLSIDCSEANQPIKSPEDVAPEAEASEDFPTTSD 1023
TV ESI + DD + K + E D+ KS E+ A+++
Sbjct: 2896 TVMESINVVVDDLSPARKKDVEE---DVRTSGDNVADAAKSGENAENSDSATDESNINQP 3066
Query: 1024 PLQKKKRIVASHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGFVSLIEPKSVDEALEDK 1083
+ RI HP+ELIIG+ + V TRS E ++S FVS IEPK+V EAL D+
Sbjct: 3067 DKRSSTRIQKMHPKELIIGDPNRGVTTRSR----EVEIVSNSCFVSKIEPKNVKEALTDE 3234
Query: 1084 GWIQAMQEELDQFTKNDVWTLMPKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYS 1143
WI AMQEEL+QF +N+VW L+P+P+G +VIGTKW+F+NK NE+G +TRNKARLVAQGY+
Sbjct: 3235 FWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYT 3414
Query: 1144 QQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVKQPPGF 1203
Q EG+D+ ETFAPVARLE+IRLL+ + L+QMDVKSAFLNGY++EEVYV+QP GF
Sbjct: 3415 QIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGF 3594
Query: 1204 EDNKHPEHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCRTYKNDILIV 1263
D HP+HVY+LKK+LYGLKQAPRAWYERL+ FL Q + +G D TLF + +++I
Sbjct: 3595 ADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIA 3774
Query: 1264 QIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTL 1323
QIYVDDI+FG + + + F + MQ+EFEMS++GEL YFLG+Q+ Q ++ Q +Y
Sbjct: 3775 QIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAK 3954
Query: 1324 ELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVH 1383
++KKF M + + +TP L K+E + V Q LYR MIGSLLYLTASRPDI ++V
Sbjct: 3955 NIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVG 4134
Query: 1384 LCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKST 1443
+CAR+Q++P+ +HLT VKRILKY+ GT++ G+MY S L G+CDAD+AG +RKST
Sbjct: 4135 VCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKST 4314
Query: 1444 SGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVPI 1503
SG C +LG+NL++W SK+QN ++LSTAEAEY++A + C+Q +WMK L++Y + + +
Sbjct: 4315 SGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTL 4494
Query: 1504 YCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDT 1549
YCDN +AI++SKNP+ HSR KHI++++HYIRD V ++L++VDT
Sbjct: 4495 YCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDT 4632
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 958 bits (2477), Expect = 0.0
Identities = 488/1007 (48%), Positives = 670/1007 (66%), Gaps = 1/1007 (0%)
Frame = +1
Query: 544 LLQTSYAAQLRHLSWYLDSGCSRHMTGTRSIFQKLTPLKSGGDVGFGGNQKGKIIGKGSI 603
++ TS A + WYLDSGCSRHMTG + + P S V FG KGKI G G +
Sbjct: 1648 VVHTSLRASAKE-DWYLDSGCSRHMTGVKEFLVNIEPC-STSYVTFGDGSKGKITGMGKL 1821
Query: 604 GDGKTPVINDVLLVEGLFHNLLSISQIADKGYDVIFNQTGCKAVSQTDGSVLFSGKRKND 663
P +N VLLV+GL NL+SISQ+ D+G++V F ++ C V+ VL G R D
Sbjct: 1822 VHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKD 1998
Query: 664 IYKIRSSELLSQKVKCLMSVNDEQWIWHRRLGHASLRKISQLSKLNLVRGLPHLKYSSEA 723
+ + + S CL S DE IWH+R GH LR + ++ VRG+P+LK
Sbjct: 1999 NCYLWTPQETSYSSTCLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGR 2178
Query: 724 LCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYELVIVDDYSRWT 783
+C CQ GK K + +T+R LELLH+DL GP++ ES+GGK+Y V+VDD+SR+T
Sbjct: 2179 ICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFT 2358
Query: 784 WVKFLRHKDETHTMFTNFITQVQKEFQSSVITVRSDHGGEFENKAFEDLFNSQGISHNFS 843
WV F+R K +T +F ++Q+E + +RSDHG EFEN F + S+GI+H FS
Sbjct: 2359 WVNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFS 2538
Query: 844 CPRTPQQNGVVERKNITLQEMARTMMQESSMAKHFWAEAINTACYIQNRISIRPILEKTP 903
TPQQNG+VERKN TLQE AR M+ + + WAEA+NTACYI NR+++R T
Sbjct: 2539 AAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTL 2718
Query: 904 YELCKGRQPDISYFHPFGSTCFMLNTKEQLGKFDSKALKCYFLGYSERSKGFRIYNIIHQ 963
YE+ KGR+P + +FH FGS C++L +EQ K D K+ FLGYS S+ +R++N +
Sbjct: 2719 YEIWKGRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTR 2898
Query: 964 TVEESIQIRFDDKLGSEKSKLFERFADLSIDCSEANQPIKSPEDVAPEAEASEDFPTTSD 1023
TV ESI + DD + K + E D+ KS E+ A ++++ D P +
Sbjct: 2899 TVMESINVVVDDLTPARKKDVEE---DVRTSGDNVADTAKSAEN-AENSDSATDEPNINQ 3066
Query: 1024 PLQKKK-RIVASHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGFVSLIEPKSVDEALED 1082
P ++ RI HP+ELIIG+ + V TRS E ++S FVS IEPK+V EAL D
Sbjct: 3067 PDKRPSIRIQKMHPKELIIGDPNRGVTTRSR----EIEIVSNSCFVSKIEPKNVKEALTD 3234
Query: 1083 KGWIQAMQEELDQFTKNDVWTLMPKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGY 1142
+ WI AMQEEL+QF +N+VW L+P+P+G +VIGTKW+F+NK NE+G +TRNKARLVAQGY
Sbjct: 3235 EFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGY 3414
Query: 1143 SQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVKQPPG 1202
+Q EG+D+ ETFAPVARLE+IRLL+ + L+QMDVKSAFLNGY++EE YV+QP G
Sbjct: 3415 TQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKG 3594
Query: 1203 FEDNKHPEHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCRTYKNDILI 1262
F D HP+HVY+LKK+LYGLKQAPRAWYERL+ FL Q + +G D TLF + +++I
Sbjct: 3595 FVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMI 3774
Query: 1263 VQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYT 1322
QIYVDDI+FG + + + F + MQ+EFEMS++GEL YFLG+Q+ Q ++ Q KY
Sbjct: 3775 AQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYA 3954
Query: 1323 LELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSV 1382
++KKF M + + +TP L K+E + V Q LYR MIGSLLYLTASRPDI ++V
Sbjct: 3955 KNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAV 4134
Query: 1383 HLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKS 1442
+CAR+Q++P+ +HL VKRILKY+ GT++ G+MY S+ L G+CDAD+AG +RKS
Sbjct: 4135 GVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSADDRKS 4314
Query: 1443 TSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVP 1502
TSG C +LG+NL++W SK+QN ++LSTAEAEY++A + C+Q +WMK L++Y + +
Sbjct: 4315 TSGGCFYLGTNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMT 4494
Query: 1503 IYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDT 1549
+YCDN +AI++SKNP+ HSR KHI++++HYIRD V ++LE+VDT
Sbjct: 4495 LYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLEHVDT 4635
>TC211311 weakly similar to UP|O24587 (O24587) Pol protein, partial (15%)
Length = 1213
Score = 207 bits (528), Expect(2) = 9e-63
Identities = 107/191 (56%), Positives = 136/191 (71%)
Frame = +3
Query: 1254 RTYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGV 1313
R K LI+ IYVDDIIFG+ + +CKEF +LM+ FE SM GELK+ LG+QI Q+
Sbjct: 468 RLKKETFLIIHIYVDDIIFGATSKRMCKEFFELMKDGFETSMKGELKFLLGLQIIQKVYG 647
Query: 1314 TYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTA 1373
+IHQ+KYT LK+F M + TPMH + I++K+E + K Y GMI SL YLT+
Sbjct: 648 IFIHQEKYTKSHLKRFRMDEAKPMATPMHRSTIIDKDEKGNHTS*KEYSGMIDSLSYLTS 827
Query: 1374 SRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADF 1433
SRPDI+F V LCARFQS P+ +H+TAVKRIL+YL GTTN L ++K SE+ L G+CD F
Sbjct: 828 SRPDIVFVVCLCARFQSYPKISHVTAVKRILRYLVGTTNHCLWFKKRSEFDLLGYCDVYF 1007
Query: 1434 AGDRVERKSTS 1444
AGD+VERKSTS
Sbjct: 1008 AGDKVERKSTS 1040
Score = 53.1 bits (126), Expect(2) = 9e-63
Identities = 26/47 (55%), Positives = 31/47 (65%)
Frame = +2
Query: 1214 KLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCRTYKNDI 1260
K +YGLKQA RAWYERLSSFL+ N F RG D LF + K ++
Sbjct: 347 KTLSCVYGLKQALRAWYERLSSFLVSNGFTRGITDPALFRKAQKGNL 487
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 221 bits (564), Expect = 1e-57
Identities = 106/152 (69%), Positives = 129/152 (84%)
Frame = -3
Query: 1088 AMQEELDQFTKNDVWTLMPKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEG 1147
AMQEEL+QF +N+VW L+ KP+ + VIGTKWVFRNKL+E G + RNKARLVA+GY+Q+EG
Sbjct: 458 AMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEEG 279
Query: 1148 IDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVKQPPGFEDNK 1207
IDY ET+APVARLE IR+L+++ N L+QMDVKSAFLNG I EEVYV+QPPGFE
Sbjct: 278 IDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIPD 99
Query: 1208 HPEHVYKLKKSLYGLKQAPRAWYERLSSFLLQ 1239
P HVYKL+K+LYGLKQAPRAWYER+S+FLL+
Sbjct: 98 KPTHVYKLQKALYGLKQAPRAWYERISNFLLE 3
>TC232995
Length = 1009
Score = 213 bits (543), Expect = 4e-55
Identities = 107/173 (61%), Positives = 126/173 (71%)
Frame = +2
Query: 1197 VKQPPGFEDNKHPEHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCRTY 1256
V+QPPGFE + P HVYKL+K+LYGLKQAPRAWYERLS+FLL+ EF RGK D TLF +
Sbjct: 2 VEQPPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRK 181
Query: 1257 KNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYI 1316
NDIL+VQIYVDDIIFGS N SLCKEFS MQ+EFEMSMMGELKYFLG+QI Q +I
Sbjct: 182 HNDILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQIKQTQ*GIFI 361
Query: 1317 HQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLL 1369
+Q KY EL+K+F M TPM C L+K+E + K YR IG ++
Sbjct: 362 NQSKYCKELIKRFGMDSAKHMSTPMSTNCYLDKDESGQSIDIKQYRDAIGEVV 520
>TC213445
Length = 705
Score = 129 bits (325), Expect(2) = 1e-48
Identities = 57/98 (58%), Positives = 78/98 (79%)
Frame = +1
Query: 1437 RVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGL 1496
+ +R+STS +CHF+GS LV+W SK+QN++ LSTAEAEY+SA + Q WM+ L DYGL
Sbjct: 400 KTDRESTSDTCHFIGSALVSWHSKKQNSVVLSTAEAEYISARSYYAQIFWMRQQLFDYGL 579
Query: 1497 SLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIR 1534
L +PI CDNT+AI+LSKN IL+SR KHIE+++H++R
Sbjct: 580 KLDHIPIRCDNTSAINLSKNHILYSRTKHIEIRHHFLR 693
Score = 84.3 bits (207), Expect(2) = 1e-48
Identities = 38/68 (55%), Positives = 51/68 (74%)
Frame = +2
Query: 1364 MIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEY 1423
MI S LYL+ SRP I+FSV +C R+Q++P+E+HL+ +KRI++YL G NLGL Y K S Y
Sbjct: 197 MIESFLYLSTSRPHIMFSVCMCVRYQANPKESHLSVIKRIMRYLLGIINLGLWYPKNSSY 376
Query: 1424 TLSGFCDA 1431
L G+ DA
Sbjct: 377 NLVGYSDA 400
>AI959950
Length = 466
Score = 166 bits (419), Expect = 1e-40
Identities = 84/132 (63%), Positives = 104/132 (78%)
Frame = -1
Query: 1086 IQAMQEELDQFTKNDVWTLMPKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQ 1145
++AMQEELDQF KN+V L+ PK V+G KW+F NKL+E G+V R KARLVA+GYSQQ
Sbjct: 397 MKAMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQ 218
Query: 1146 EGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVKQPPGFED 1205
EGIDY +TFA VARLE I +L+SF+ N+ L+QMDVKSAFLNG I +EVYV+QPPGFE+
Sbjct: 217 EGIDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGFEN 38
Query: 1206 NKHPEHVYKLKK 1217
+HV+KL K
Sbjct: 37 ETLHQHVFKLNK 2
>TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein, partial
(7%)
Length = 804
Score = 162 bits (409), Expect = 1e-39
Identities = 86/190 (45%), Positives = 124/190 (65%), Gaps = 4/190 (2%)
Frame = +1
Query: 1361 YRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMY--- 1417
+R +IGSL YL SRP+I F+V L +RF PR +H+ A KR+L+ +KGT G+++
Sbjct: 22 FRRLIGSLRYLCNSRPNICFAVSLISRFMKRPRLSHMQAAKRVLRLIKGTIGSGVLFPFK 201
Query: 1418 RKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSA 1477
K+ + L G+ D+D+ D + KST G V SSK+Q+ IALST EAEYV+A
Sbjct: 202 AKSGKPDLLGYTDSDWKRDPEQEKSTGGYLFMYNDAPVA*SSKKQDVIALSTCEAEYVAA 381
Query: 1478 ATCCTQTIWMKNHLEDYGLSLKK-VPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDH 1536
+ Q +WM N LE+ L +K V + DN +AI+L+K+P LH R+KHIE+++HYIRD
Sbjct: 382 SLGACQAVWMMNLLEELKLRERKPVNLLIDNKSAINLAKHPTLHGRSKHIELRFHYIRDQ 561
Query: 1537 VQKGTLSLEY 1546
V KG +++EY
Sbjct: 562 VSKGNVTVEY 591
>BM143109
Length = 415
Score = 155 bits (393), Expect = 1e-37
Identities = 80/135 (59%), Positives = 96/135 (70%)
Frame = +1
Query: 1199 QPPGFEDNKHPEHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCRTYKN 1258
QPP ++++ P HV+KLKK LYGLKQA RAWYE LS FLL F +GK D LF N
Sbjct: 4 QPPVRKNSEKPNHVFKLKKVLYGLKQALRAWYELLSKFLLDKGFSKGKVDTNLFI*KKLN 183
Query: 1259 DILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQ 1318
DIL+VQIYVDDIIFGS N SLCK+FS+ MQ EFEMSMM EL +FLG+QI Q +I Q
Sbjct: 184 DILLVQIYVDDIIFGSTNDSLCKKFSQDMQNEFEMSMMRELNFFLGLQIKQTKNGIFISQ 363
Query: 1319 KKYTLELLKKFNMSD 1333
KY +L+ +F M +
Sbjct: 364 SKYCKDLIHRFGMEN 408
>CO983516
Length = 724
Score = 152 bits (385), Expect = 8e-37
Identities = 74/120 (61%), Positives = 92/120 (76%)
Frame = +2
Query: 1154 FAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVKQPPGFEDNKHPEHVY 1213
F PVARLE+IRLL+ + L+QMDVKSAFLNGY++EEVYV+QP GF D HP+HVY
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 1214 KLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCRTYKNDILIVQIYVDDIIFG 1273
+LKK+LYGLKQAPRAWYERL+ L Q + +G D TLF + +++I QIYVDDI+FG
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFG 724
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 149 bits (377), Expect = 7e-36
Identities = 70/111 (63%), Positives = 91/111 (81%)
Frame = +3
Query: 1072 EPKSVDEALEDKGWIQAMQEELDQFTKNDVWTLMPKPKGFHVIGTKWVFRNKLNEKGEVT 1131
EPK++ EA+ D WI MQEEL+QF +N+VW L+ KP+ + VIGTKWVFRNKL+E G +
Sbjct: 3 EPKNIKEAIVDDNWIIVMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIII 182
Query: 1132 RNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDV 1182
RNKARLVA+GY+Q+EGIDY ET+APVARLEAIR+L++++ N L+QMDV
Sbjct: 183 RNKARLVAKGYNQEEGIDYEETYAPVARLEAIRMLLAYASIMNFKLYQMDV 335
>AI855982
Length = 484
Score = 142 bits (358), Expect = 1e-33
Identities = 79/165 (47%), Positives = 106/165 (63%)
Frame = +2
Query: 1036 PEELIIGNKDAPVRTRSMLKPSEETLLSLKGFVSLIEPKSVDEALEDKGWIQAMQEELDQ 1095
P + IIG+ V TR LK L + FVS+IEPK++ EA+ D WI AMQEEL+Q
Sbjct: 2 PLDNIIGDISKGVTTRHSLKD----LCNNMAFVSMIEPKNIKEAIVDDNWIIAMQEELNQ 169
Query: 1096 FTKNDVWTLMPKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFA 1155
F +N+VW L+ KP + VI TKWVFRNKL+E + +KARLVA+GY+Q +G+DY T+A
Sbjct: 170 FERNNVWKLVEKPDNYPVI*TKWVFRNKLDEHRIIIIHKARLVAEGYNQVDGLDYEHTYA 349
Query: 1156 PVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVKQP 1200
+ARL I + +S+ N L+ SA L+G + EVYV QP
Sbjct: 350 SIARL*VIIMPLSYVYIMNSTLYHYACVSALLHGLLLHEVYVDQP 484
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 131 bits (330), Expect = 2e-30
Identities = 63/130 (48%), Positives = 91/130 (69%)
Frame = -2
Query: 1105 MPKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIR 1164
+P P G +G +WV+ K+ GEV R KARLVA+GY+Q GIDY +TF+PVA+L +R
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 1165 LLISFSVNHNIILHQMDVKSAFLNGYISEEVYVKQPPGFEDNKHPEHVYKLKKSLYGLKQ 1224
L ++ + + LHQ+D+K+AFL+G + E++Y++QPPGF V KL +SLYGLKQ
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 1225 APRAWYERLS 1234
+PRAW+ + S
Sbjct: 46 SPRAWFGKFS 17
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 123 bits (309), Expect = 5e-28
Identities = 57/132 (43%), Positives = 87/132 (65%)
Frame = +3
Query: 1069 SLIEPKSVDEALEDKGWIQAMQEELDQFTKNDVWTLMPKPKGFHVIGTKWVFRNKLNEKG 1128
SL P ++ EAL+ GW QAM +E+ N W L+P P G +G +WV+ K+ G
Sbjct: 9 SLTVPSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGPNG 188
Query: 1129 EVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLN 1188
+V R KARLVA+GY+Q GI+Y +TF+PV L +RL ++ + + LHQ+D+K+AFL+
Sbjct: 189 KVDRLKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAFLH 368
Query: 1189 GYISEEVYVKQP 1200
G + E++Y++QP
Sbjct: 369 GDLEEDIYMEQP 404
>BG508993
Length = 374
Score = 120 bits (302), Expect = 4e-27
Identities = 58/123 (47%), Positives = 80/123 (64%), Gaps = 1/123 (0%)
Frame = +1
Query: 1408 KGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIAL 1467
KGT + GL Y ++ Y L GFCD+DFAGD +RKST+G F+G + TWSSK+Q + L
Sbjct: 4 KGTIDFGLFYSPSNNYKLVGFCDSDFAGDVDDRKSTTGFVFFMGDCVFTWSSKKQGIVTL 183
Query: 1468 STAEAEYVSAATCCTQTIWMKNHLEDYGLSLKK-VPIYCDNTAAISLSKNPILHSRAKHI 1526
T EAEYV+A +C IW++ LE+ L K+ IY DN +A L+KN + H R+KHI
Sbjct: 184 FTCEAEYVAATSCTCHAIWLRRLLEELQLLQKESTKIYVDNRSAQELAKNSVFHERSKHI 363
Query: 1527 EVK 1529
+ +
Sbjct: 364 DTR 372
>CO982036
Length = 674
Score = 120 bits (302), Expect = 4e-27
Identities = 72/212 (33%), Positives = 117/212 (54%), Gaps = 5/212 (2%)
Frame = -2
Query: 1256 YKNDILIVQ--IYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGV 1313
YK IL V +YVD II GS+ +L + + + + F + ++G+L YF+ I++ P +
Sbjct: 673 YKTHILTVYLLVYVDIIITGSSC-TLIQNLTSKLNSSFPLKLLGKLDYFVEIEVKSMPDL 497
Query: 1314 TYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTA 1373
+ + E+ + +PM TC L K + YR ++G+L Y T
Sbjct: 496 LF-SLRTSIFEIFCRKPR*QAQPISSPMTTTCKLSKSDSDLFSGPTFYRSVVGALQYTTV 320
Query: 1374 SRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYR---KTSEYTLSGFCD 1430
RP+I F+V+ +F S+P ++H T VKRIL+YLKG+ + GL + + + GFCD
Sbjct: 319 IRPEISFAVNKVCQFMSNPLDSHWTEVKRILRYLKGSLSYGL*LKPAISSQPLPIRGFCD 140
Query: 1431 ADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQ 1462
AD+A +++STSG+ FLG NL++W +Q
Sbjct: 139 ADWASAVDDKRSTSGAAVFLGPNLISWWXXKQ 44
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment), partial
(30%)
Length = 687
Score = 120 bits (300), Expect = 6e-27
Identities = 54/124 (43%), Positives = 84/124 (67%), Gaps = 1/124 (0%)
Frame = +2
Query: 1425 LSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQT 1484
LSG+CDAD+AG ++R+STSG C F+G NLV+W SK+Q +A S+AEAEY S A +
Sbjct: 17 LSGYCDADWAGCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMAMVTCEL 196
Query: 1485 IWMKNHLEDYGLSLK-KVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLS 1543
+W+K L++ + ++ +YCDN AA+ ++ NP+ H R KHIE+ H+IR+ + +
Sbjct: 197 MWIKQFLQELRFCEELQMKLYCDNQAALHIASNPVFHERTKHIEIDCHFIREKLLSKEIV 376
Query: 1544 LEYV 1547
E++
Sbjct: 377 TEFI 388
>BU549979
Length = 615
Score = 118 bits (296), Expect = 2e-26
Identities = 59/166 (35%), Positives = 101/166 (60%), Gaps = 3/166 (1%)
Frame = -1
Query: 1387 RFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGS 1446
R+QS+P H K++++YL+GT + LMY++T+ + G+ D+DFAG R+STSG
Sbjct: 606 RYQSNPGIDHWKTAKKVMRYLQGTKDYMLMYKQTNCLEVIGYSDSDFAGCVDSRRSTSGY 427
Query: 1447 CHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGL---SLKKVPI 1503
L +V+W S +Q IA ST E E+V + +W+K+ + + + + +
Sbjct: 426 IFMLADGVVSWRSSKQTLIATSTMEVEFVPCFEATSHGVWLKSFMSSLRVVDSISRPLKL 247
Query: 1504 YCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDT 1549
YCDN AA+ ++KN +R+KHI++KY IR+ V++ + +E+V+T
Sbjct: 246 YCDNFAAVFMAKNNKSGNRSKHIDIKYLVIRERVKEKKVVIEHVNT 109
>BI425121
Length = 412
Score = 113 bits (282), Expect = 7e-25
Identities = 68/152 (44%), Positives = 89/152 (57%)
Frame = +3
Query: 765 SIGGKKYELVIVDDYSRWTWVKFLRHKDETHTMFTNFITQVQKEFQSSVITVRSDHGGEF 824
S+G KKYE + VDDYSR+TWV FL HK E+ F +++K F ++ V +
Sbjct: 6 SLGCKKYEFLTVDDYSRYTWVYFLAHKHESLRYFIRGF-KMKKVFVFLLLEVTMELS--L 176
Query: 825 ENKAFEDLFNSQGISHNFSCPRTPQQNGVVERKNITLQEMARTMMQESSMAKHFWAEAIN 884
+ GI HN S PRTPQ+N VVERKN TLQE ART+ +
Sbjct: 177 RILS*NHFCERNGIFHNLS*PRTPQENRVVERKNRTLQEKARTI--------------LF 314
Query: 885 TACYIQNRISIRPILEKTPYELCKGRQPDISY 916
T C++QN+I IRP+++KTPYEL KGR+ ISY
Sbjct: 315 TTCFVQNKILIRPMIKKTPYELWKGRRHIISY 410
>BI321712
Length = 399
Score = 110 bits (275), Expect = 5e-24
Identities = 58/124 (46%), Positives = 77/124 (61%)
Frame = -3
Query: 1265 IYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLE 1324
+YVDD+IF NPS+ +EF K M EFEM+ MG + Y+LGI++ Q +I Q+ Y E
Sbjct: 379 LYVDDLIFTGNNPSMFEEFKKDMSNEFEMTDMGLMAYYLGIEVKQEDKGIFITQEGYAKE 200
Query: 1325 LLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHL 1384
+LKKF M D N TPM L K E V LY+ +IGSL YLT +RPDIL+ V +
Sbjct: 199 VLKKFKMDDANPVGTPMECGSKLSKHEKGENVDPTLYKSLIGSLRYLTCTRPDILYVVGV 20
Query: 1385 CARF 1388
+R+
Sbjct: 19 VSRY 8
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.330 0.140 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 68,645,155
Number of Sequences: 63676
Number of extensions: 969736
Number of successful extensions: 6374
Number of sequences better than 10.0: 138
Number of HSP's better than 10.0 without gapping: 6226
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6324
length of query: 1549
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1439
effective length of database: 5,635,272
effective search space: 8109156408
effective search space used: 8109156408
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0240.16


