

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
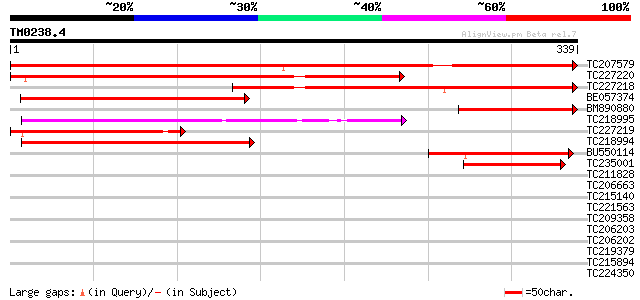
Query= TM0238.4
(339 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207579 similar to GB|AAN15531.1|23198008|BT000212 expressed pr... 449 e-126
TC227220 similar to GB|AAN15531.1|23198008|BT000212 expressed pr... 351 2e-97
TC227218 similar to GB|AAN15531.1|23198008|BT000212 expressed pr... 335 1e-92
BE057374 weakly similar to GP|14517356|gb| AT4g15540/dl3810w {Ar... 151 4e-37
BM890880 similar to GP|14517356|gb| AT4g15540/dl3810w {Arabidops... 135 2e-32
TC218995 similar to GB|AAD34703.1|4966372|F3O9 ESTs gb|N38586 an... 133 1e-31
TC227219 similar to GB|AAN15531.1|23198008|BT000212 expressed pr... 128 4e-30
TC218994 weakly similar to UP|Q9SGT5 (Q9SGT5) T6H22.12 protein, ... 120 7e-28
BU550114 similar to GP|4966372|gb|A ESTs gb|N38586 and gb|N38613... 103 1e-22
TC235001 homologue to GB|AAD34703.1|4966372|F3O9 ESTs gb|N38586 ... 97 1e-20
TC211828 similar to UP|RPIA_ARATH (Q9ZU38) Probable-ribose 5-pho... 39 0.003
TC206663 weakly similar to GB|AAM65353.1|21655283|AY103301 At1g2... 38 0.006
TC215140 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 37 0.010
TC221563 similar to UP|O23702 (O23702) Dehydrogenase, partial (21%) 37 0.010
TC209358 similar to UP|P92987 (P92987) Myosin heavy chain-like p... 37 0.013
TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragme... 37 0.017
TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragme... 37 0.017
TC219379 similar to UP|Q941E8 (Q941E8) AT3g04880/T9J14_17, parti... 36 0.022
TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase ... 36 0.022
TC224350 similar to UP|Q872T5 (Q872T5) Probable 60S large subuni... 35 0.037
>TC207579 similar to GB|AAN15531.1|23198008|BT000212 expressed protein
{Arabidopsis thaliana;} , partial (57%)
Length = 1249
Score = 449 bits (1154), Expect = e-126
Identities = 237/342 (69%), Positives = 280/342 (81%), Gaps = 3/342 (0%)
Frame = +2
Query: 1 MAAESGDLNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEK 60
+ +ES DLP++L+ VLPSDP+EQLD+ARKITS+ALS RV+AL+SESS LRA +A++
Sbjct: 17 LVSESSGSKVDLPEELLNVLPSDPYEQLDVARKITSVALSTRVDALQSESSALRAELADR 196
Query: 61 DHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFR 120
+ LIAELQSQ++S+DA LS AD L RA+QDKE+L+ ENASLSNTVRKL RDVSKLE FR
Sbjct: 197 NRLIAELQSQVESIDAALSEAADKLARADQDKENLLKENASLSNTVRKLTRDVSKLETFR 376
Query: 121 KTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEAS--LPPSVSSSVPSISDTG 178
KTLM+SL+E++D S G D AK+ SQ+S TSTSQFGD++AS L SS + SD G
Sbjct: 377 KTLMKSLREDEDTSEGTADTAAKLHSQASFTSTSQFGDDDASSTLSSRTSSMRINTSDMG 556
Query: 179 NSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRT-SKPVSPRRHS 237
N AED ESD + R ++LLLASQ++TPR+TPPGSPPS+SA VSPTRT SKPVSPRRH+
Sbjct: 557 NYLAEDRESDGSKSRASHNLLLASQTSTPRITPPGSPPSMSALVSPTRTSSKPVSPRRHA 736
Query: 238 ISFATSRGMHDDRTSVFSSMSSSDSGAGSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLAN 297
ISF+TSRGM DDR+SV GSQTGRTRVDGKEFFRQVR+RLSYEQFGAFLAN
Sbjct: 737 ISFSTSRGMFDDRSSV-----------GSQTGRTRVDGKEFFRQVRSRLSYEQFGAFLAN 883
Query: 298 VKELNSHKQTREVTLQKADEIFGPENKDLYNIFEGLITRNVH 339
VKELNSHKQT+E TLQKA+E+FGPENKDLY IFEGLITRNVH
Sbjct: 884 VKELNSHKQTKEETLQKANELFGPENKDLYTIFEGLITRNVH 1009
>TC227220 similar to GB|AAN15531.1|23198008|BT000212 expressed protein
{Arabidopsis thaliana;} , partial (43%)
Length = 927
Score = 351 bits (901), Expect = 2e-97
Identities = 186/238 (78%), Positives = 209/238 (87%), Gaps = 2/238 (0%)
Frame = +3
Query: 1 MAAESGDL--NFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIA 58
MAAESG NFDLP+++VQVLPSDPF+QLD+ARKITSIALS RVN LESE S LRA+IA
Sbjct: 231 MAAESGGTPTNFDLPEEVVQVLPSDPFQQLDVARKITSIALSTRVNTLESELSSLRAQIA 410
Query: 59 EKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEV 118
+KD+LIA+LQSQLDSLDA+LS AD L + EQDKESL+ ENASLSNTV+KLNRDVSKLEV
Sbjct: 411 DKDNLIADLQSQLDSLDASLSQIADKLLQTEQDKESLLQENASLSNTVKKLNRDVSKLEV 590
Query: 119 FRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTG 178
FRKTLM+SLQE+DDNSG PD VAK+QSQ+SLTSTSQ GDNEASLPP++SSS TG
Sbjct: 591 FRKTLMQSLQEDDDNSGATPDTVAKIQSQASLTSTSQIGDNEASLPPAISSS------TG 752
Query: 179 NSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRH 236
NSFA+D ESDAIRPRV +LLLASQ +TPR+TPPGSPP LSASVSPTRTSKPVSPRRH
Sbjct: 753 NSFADDQESDAIRPRVSQNLLLASQGSTPRITPPGSPPILSASVSPTRTSKPVSPRRH 926
>TC227218 similar to GB|AAN15531.1|23198008|BT000212 expressed protein
{Arabidopsis thaliana;} , partial (42%)
Length = 787
Score = 335 bits (860), Expect = 1e-92
Identities = 172/213 (80%), Positives = 190/213 (88%), Gaps = 7/213 (3%)
Frame = +1
Query: 134 SGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPR 193
+G APD+VAK+QSQ+SLTSTSQ GDN+ SLPPSVSSS TGNSFA+DHESDAIRPR
Sbjct: 1 AGAAPDIVAKIQSQASLTSTSQIGDNDVSLPPSVSSS------TGNSFAKDHESDAIRPR 162
Query: 194 VPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSV 253
V +LLLASQ +TPR+TPPGSPPSLSASVSPTRTSKPVSP+RHSISFAT+RGM+DDR+S+
Sbjct: 163 VSQNLLLASQGSTPRITPPGSPPSLSASVSPTRTSKPVSPQRHSISFATTRGMYDDRSSM 342
Query: 254 FSSMS-------SSDSGAGSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLANVKELNSHKQ 306
FSSMS SSD+G GSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLANVKELNSHKQ
Sbjct: 343 FSSMSLTHGSISSSDAGTGSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLANVKELNSHKQ 522
Query: 307 TREVTLQKADEIFGPENKDLYNIFEGLITRNVH 339
T+E TL+KADEIFGPENKDLY IFEGLI RN+H
Sbjct: 523 TKEETLRKADEIFGPENKDLYTIFEGLINRNLH 621
>BE057374 weakly similar to GP|14517356|gb| AT4g15540/dl3810w {Arabidopsis
thaliana}, partial (13%)
Length = 412
Score = 151 bits (382), Expect = 4e-37
Identities = 84/137 (61%), Positives = 105/137 (76%)
Frame = +2
Query: 7 DLNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAE 66
+++FDL +++ VLPSDPF+Q+D+ RKIT IALS RVN L SE S L A+I + D+LI +
Sbjct: 2 NMDFDLTEEVSLVLPSDPFQQVDVDRKITYIALSTRVNTL*SELSSLAAQITD*DNLIDD 181
Query: 67 LQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRS 126
LQ LDSLDA LS L + EQDKESL+ ENASL ++++KLNRDV KLEV +TLM+S
Sbjct: 182 LQV*LDSLDAYLSQIGATLLQTEQDKESLLQENASLYDSMKKLNRDVYKLEVIIETLMQS 361
Query: 127 LQEEDDNSGGAPDMVAK 143
LQE DDNSGGA D VAK
Sbjct: 362 LQEVDDNSGGAADTVAK 412
>BM890880 similar to GP|14517356|gb| AT4g15540/dl3810w {Arabidopsis
thaliana}, partial (20%)
Length = 421
Score = 135 bits (341), Expect = 2e-32
Identities = 65/71 (91%), Positives = 68/71 (95%)
Frame = +1
Query: 269 GRTRVDGKEFFRQVRNRLSYEQFGAFLANVKELNSHKQTREVTLQKADEIFGPENKDLYN 328
GRTRVDGKEFFRQVRNRLSYEQFGAFLANVKELNSHKQT+E TL+KADEIFGPENKDLY
Sbjct: 169 GRTRVDGKEFFRQVRNRLSYEQFGAFLANVKELNSHKQTKEETLRKADEIFGPENKDLYT 348
Query: 329 IFEGLITRNVH 339
IFEGLI RN+H
Sbjct: 349 IFEGLINRNLH 381
>TC218995 similar to GB|AAD34703.1|4966372|F3O9 ESTs gb|N38586 and gb|N38613
come from this gene. {Arabidopsis thaliana;} , partial
(33%)
Length = 759
Score = 133 bits (335), Expect = 1e-31
Identities = 87/230 (37%), Positives = 133/230 (57%)
Frame = +3
Query: 8 LNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAEL 67
++FDLPD+++ V+P+DP++QLDLARKITS+A+++RV++LES++S LR ++ EKD +I +L
Sbjct: 66 VDFDLPDEILSVIPTDPYQQLDLARKITSMAIASRVSSLESDASRLRQKLLEKDRIILDL 245
Query: 68 QSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSL 127
+ +L SL + L+ A + L E L+ TV+KL+RD +KLE F+K LM+SL
Sbjct: 246 EDRLSSLTRASHQSDSTLNTALNENIKLTKERDQLAATVKKLSRDFAKLETFKKQLMQSL 425
Query: 128 QEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHES 187
DDN+ A QS + D+ + S + P +D G + E
Sbjct: 426 --TDDNALHAETTDIGTCDQSVPKAYPDKDDDRSGNMAHHSYNGP--ADVGKTNDEASRY 593
Query: 188 DAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHS 237
R +SL + TPRLTP G+P +S + SP S VSP++ S
Sbjct: 594 SGQR----FSL---TPYITPRLTPTGTPKVISTAGSPRGYSAAVSPKKTS 722
>TC227219 similar to GB|AAN15531.1|23198008|BT000212 expressed protein
{Arabidopsis thaliana;} , partial (13%)
Length = 421
Score = 128 bits (321), Expect = 4e-30
Identities = 72/109 (66%), Positives = 88/109 (80%), Gaps = 4/109 (3%)
Frame = +1
Query: 1 MAAESG----DLNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRAR 56
MA ESG +LNFDLP+++VQVLPSDPF+QLD+ARKITSIALS RVN LES+ S LRA+
Sbjct: 91 MAGESGGSATNLNFDLPEEVVQVLPSDPFQQLDVARKITSIALSTRVNTLESDLSSLRAQ 270
Query: 57 IAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNT 105
IA+KD+LIA+LQSQLDSLDA+LS A L + EQDK ++ SLS+T
Sbjct: 271 IADKDNLIADLQSQLDSLDASLSQIAATLFQTEQDK--VLTNFHSLSHT 411
>TC218994 weakly similar to UP|Q9SGT5 (Q9SGT5) T6H22.12 protein, partial
(35%)
Length = 538
Score = 120 bits (302), Expect = 7e-28
Identities = 60/139 (43%), Positives = 97/139 (69%)
Frame = +2
Query: 8 LNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAEL 67
++FDLPD+++ V+P+DP++QLDLARKITS+A+++RV++LES++S LR ++ EKD +I +L
Sbjct: 92 VDFDLPDEILSVIPTDPYQQLDLARKITSMAIASRVSSLESDASRLRQKLLEKDRIILDL 271
Query: 68 QSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSL 127
+ +L SL T L+ A + L E L+ TV+KL+RD +KLE F+K LM+SL
Sbjct: 272 EDRLSSLTRASHQTDSTLNTALNENIKLSKERDQLAATVKKLSRDFAKLETFKKQLMQSL 451
Query: 128 QEEDDNSGGAPDMVAKVQS 146
+++ + G ++ QS
Sbjct: 452 TDDNASHAGTINIRTYYQS 508
>BU550114 similar to GP|4966372|gb|A ESTs gb|N38586 and gb|N38613 come from
this gene. {Arabidopsis thaliana}, partial (28%)
Length = 605
Score = 103 bits (256), Expect = 1e-22
Identities = 53/93 (56%), Positives = 66/93 (69%), Gaps = 6/93 (6%)
Frame = -2
Query: 251 TSVFSSMSSSDSGAGSQTGRT------RVDGKEFFRQVRNRLSYEQFGAFLANVKELNSH 304
+S +SS S + GR+ ++DGKE RQ R+RLSYEQF AFLAN+KELN+
Sbjct: 571 SSWYSSSQQSSAANSPPRGRSLPVRTPKIDGKEXXRQARSRLSYEQFSAFLANIKELNAQ 392
Query: 305 KQTREVTLQKADEIFGPENKDLYNIFEGLITRN 337
KQTRE TL+KADEIFG +NKDLY F+GL+ RN
Sbjct: 391 KQTREETLRKADEIFGSDNKDLYLSFQGLLNRN 293
>TC235001 homologue to GB|AAD34703.1|4966372|F3O9 ESTs gb|N38586 and
gb|N38613 come from this gene. {Arabidopsis thaliana;} ,
partial (20%)
Length = 482
Score = 97.1 bits (240), Expect = 1e-20
Identities = 46/61 (75%), Positives = 53/61 (86%)
Frame = +3
Query: 272 RVDGKEFFRQVRNRLSYEQFGAFLANVKELNSHKQTREVTLQKADEIFGPENKDLYNIFE 331
++DGKEFFRQ R+RLSYEQF AFLAN+KELN+ KQTRE TL+KADEIFG +NKDLY F
Sbjct: 60 KIDGKEFFRQARSRLSYEQFSAFLANIKELNAQKQTREETLRKADEIFGSDNKDLYLSF* 239
Query: 332 G 332
G
Sbjct: 240 G 242
>TC211828 similar to UP|RPIA_ARATH (Q9ZU38) Probable-ribose 5-phosphate
isomerase (Phosphoriboisomerase) , partial (47%)
Length = 729
Score = 38.9 bits (89), Expect = 0.003
Identities = 35/126 (27%), Positives = 56/126 (43%)
Frame = +2
Query: 137 APDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPY 196
+P ++ + +STS + AS P + S+ PS + +S E+ ++ + P P
Sbjct: 359 SPKTTSRKSPPTRPSSTSSPAWSSASAPAPLPST-PSTASASSSAKENSKTSSESPPPPK 535
Query: 197 SLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSS 256
S S +P +PPS+S S +PTR+ + R + A S R SS
Sbjct: 536 PTTRPSPSASPSPISTPTPPSISPSTAPTRSIPSSTSSR--AAAAPSFEKRWSRAPARSS 709
Query: 257 MSSSDS 262
SSS S
Sbjct: 710 SSSSTS 727
>TC206663 weakly similar to GB|AAM65353.1|21655283|AY103301
At1g24265/At1g24265 {Arabidopsis thaliana;} , partial
(23%)
Length = 1540
Score = 38.1 bits (87), Expect = 0.006
Identities = 48/165 (29%), Positives = 63/165 (38%), Gaps = 16/165 (9%)
Frame = +1
Query: 35 TSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKES 94
T LSAR+N L+ E A I +Q + D T+S + + H A ES
Sbjct: 670 TKKKLSARINGLDKNLEECAAITESTREDIYVIQRKAD----TISEDSKSFHVAVHVLES 837
Query: 95 LINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEE----DDNSGGAPDMVAKVQSQSSL 150
I E + V+ L F KTL S E +S P + S SS
Sbjct: 838 KIKE---IEEKQVATIEGVNMLCQFTKTLENSRSTEYIQASSSSSSRPALELPPVSPSSR 1008
Query: 151 TSTSQFG------------DNEASLPPSVSSSVPSISDTGNSFAE 183
S S ASLPP++S+ PS S +G SF E
Sbjct: 1009GSQSGSARLSLEPPSVTPSSRTASLPPTLSTDPPSPSISGGSFQE 1143
>TC215140 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(91%)
Length = 1885
Score = 37.4 bits (85), Expect = 0.010
Identities = 37/128 (28%), Positives = 56/128 (42%), Gaps = 8/128 (6%)
Frame = +1
Query: 141 VAKVQSQSSLTSTSQFGDNEA---SLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYS 197
+A+ S S T+TS GD+ A S PP SSSVP +S + + P+
Sbjct: 76 LARA*S*SDTTATS--GDSTAPNSSTPPPNSSSVPLS*SHSSSSSSSPSTTLPSPQTSPP 249
Query: 198 LLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPV-----SPRRHSISFATSRGMHDDRTS 252
++ + T PP SPP S S +P + P SP + S AT+ + +
Sbjct: 250 TATSTPTPTSSPPPPPSPPGRSRSATPPPLAAPTASPSSSPAPLASSAATAPLPSKNAAT 429
Query: 253 VFSSMSSS 260
FS ++S
Sbjct: 430 AFSGSTTS 453
>TC221563 similar to UP|O23702 (O23702) Dehydrogenase, partial (21%)
Length = 486
Score = 37.4 bits (85), Expect = 0.010
Identities = 37/123 (30%), Positives = 55/123 (44%), Gaps = 6/123 (4%)
Frame = +2
Query: 148 SSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTP 207
+S T+ S + AS P ++ S + S T S S P P AS TT
Sbjct: 113 ASRTAPSNSNPSPASPPSNMFPS--AASPTARSSPPPPCSSTPSPTSPAPPSAASAPTTS 286
Query: 208 RL-----TPPGSPPSLSASVSPTRTSKPVSPRR-HSISFATSRGMHDDRTSVFSSMSSSD 261
T P +PPS S S + TS P +PRR ++S+ +S + RTS ++ S
Sbjct: 287 SSALGPPTAPSTPPSPLTSASASSTSTPRAPRRSRTLSWRSSSASYAARTSSPATRSPPP 466
Query: 262 SGA 264
+G+
Sbjct: 467 AGS 475
>TC209358 similar to UP|P92987 (P92987) Myosin heavy chain-like protein,
partial (17%)
Length = 740
Score = 37.0 bits (84), Expect = 0.013
Identities = 36/132 (27%), Positives = 60/132 (45%), Gaps = 7/132 (5%)
Frame = +1
Query: 52 ELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESLIN-ENASLSNTVRKLN 110
EL+ ++ EKD L+ ++ D ++ +A D L +KESL+ LS+ KL
Sbjct: 22 ELQRKLLEKDELLKSAENTRDQMN-VFNAKLDELKHQASEKESLLKYTQQQLSDAKIKLA 198
Query: 111 RDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQS------SLTSTSQFGDNEASLP 164
+ LE + M S ++ D M A + S + S T T+++ D + +
Sbjct: 199 DKQAALEKIQWEAMTSNKKVDKLQDELGSMQADITSFTLLLEGLSKTDTAKYTD-DYDVK 375
Query: 165 PSVSSSVPSISD 176
P S +PSI D
Sbjct: 376 PYDFSHLPSIDD 411
>TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragment) , partial
(96%)
Length = 2604
Score = 36.6 bits (83), Expect = 0.017
Identities = 44/147 (29%), Positives = 63/147 (41%), Gaps = 10/147 (6%)
Frame = +1
Query: 149 SLTSTSQFGDNEASLP--PSVSSSVPSISDTGNSFAEDHESDAIRPR-VPYSLLLASQST 205
S TS S + A P P S + PS S T S++ S + P+ YS LA +
Sbjct: 1510 SPTSPSYSPTSPAYSPTSPGYSPTSPSYSPTSPSYSPT--SPSYNPQSAKYSPSLAYSPS 1683
Query: 206 TPRLTP--PGSPPS-----LSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMS 258
+PRL+P P SP S S S SPT S S +S S + G+ D + S
Sbjct: 1684 SPRLSPSSPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSGVSPDYSPSSPQFS 1863
Query: 259 SSDSGAGSQTGRTRVDGKEFFRQVRNR 285
S + SQ G + ++ Q ++
Sbjct: 1864 PSTGYSPSQPGYSPSSTSQYTPQTSDK 1944
>TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragment) , complete
Length = 1826
Score = 36.6 bits (83), Expect = 0.017
Identities = 44/147 (29%), Positives = 63/147 (41%), Gaps = 10/147 (6%)
Frame = +3
Query: 149 SLTSTSQFGDNEASLP--PSVSSSVPSISDTGNSFAEDHESDAIRPR-VPYSLLLASQST 205
S TS S + A P P S + PS S T S++ S + P+ YS LA +
Sbjct: 1032 SPTSPSYSPTSPAYSPTSPGYSPTSPSYSPTSPSYSPT--SPSYNPQSAKYSPSLAYSPS 1205
Query: 206 TPRLTP--PGSPPS-----LSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMS 258
+PRL+P P SP S S S SPT S S +S S + G+ D + S
Sbjct: 1206 SPRLSPSSPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSGVSPDYSPSSPQFS 1385
Query: 259 SSDSGAGSQTGRTRVDGKEFFRQVRNR 285
S + SQ G + ++ Q ++
Sbjct: 1386 PSTGYSPSQPGYSPSSTSQYTPQTSDK 1466
>TC219379 similar to UP|Q941E8 (Q941E8) AT3g04880/T9J14_17, partial (81%)
Length = 1003
Score = 36.2 bits (82), Expect = 0.022
Identities = 32/106 (30%), Positives = 49/106 (46%), Gaps = 8/106 (7%)
Frame = +3
Query: 138 PDMVAKVQSQ--SSLTSTSQFGDNEASLP-PSVSSSVPSISDTGN-----SFAEDHESDA 189
P M + + S+ SSLT ++ S P P+ S++++ N S A + A
Sbjct: 99 PPMTSALLSKTPSSLTCALSTSKSKISEPLPTTPPEPRSVAESTNPPPPTSVASSPAAPA 278
Query: 190 IRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRR 235
P P S ST PR +PP +P + + S +PT + P S RR
Sbjct: 279 --PASPSSPTNIPASTPPRASPPPTPSTPAPSTTPTSSPSPASTRR 410
>TC215894 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase , complete
Length = 1675
Score = 36.2 bits (82), Expect = 0.022
Identities = 38/120 (31%), Positives = 49/120 (40%), Gaps = 3/120 (2%)
Frame = +1
Query: 138 PDMVAKVQSQSSLTSTSQFGDNE-ASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPY 196
P A +S SS +T ++ +SL S +SS S +S A S P P
Sbjct: 253 PRPSATTRSPSSAPATWEWPSRRPSSLRTSPTSSSSSTPTPTSSAARCSTSSTPPPSSP- 429
Query: 197 SLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFA--TSRGMHDDRTSVF 254
PR TPP +PPS A PT S P +P R S A +SRG +F
Sbjct: 430 ---------APRSTPPPTPPSPPA---PTSASSPPAPARSPASHASTSSRGTSPSSAPLF 573
Score = 30.8 bits (68), Expect = 0.92
Identities = 26/86 (30%), Positives = 37/86 (42%)
Frame = +1
Query: 142 AKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLA 201
A +S +S STS G + +S P SSV + +SF S
Sbjct: 499 APARSPASHASTSSRGTSPSSAPLFPLSSVTPPTQLSSSFPTPSTS-------------- 636
Query: 202 SQSTTPRLTPPGSPPSLSASVSPTRT 227
S T R + P SPP+ S++ +PT T
Sbjct: 637 --SPTSRGSSPASPPTASSAPAPTWT 708
>TC224350 similar to UP|Q872T5 (Q872T5) Probable 60S large subunit ribosomal
protein, complete
Length = 540
Score = 35.4 bits (80), Expect = 0.037
Identities = 46/149 (30%), Positives = 60/149 (39%), Gaps = 9/149 (6%)
Frame = +1
Query: 139 DMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSV---PSISDTGNSFAEDHESDAIRPRVP 195
D+ +S SS ST+ + P S +SSV PS S RP
Sbjct: 94 DVTPARRSSSSCPSTTAPRATPSPTPLSPASSVTLPPSPSACPRP---------ARPSAA 246
Query: 196 YSLLLASQSTTPRLTPPGSPPSLSASVSPT------RTSKPVSPRRHSISFATSRGMHDD 249
S L + STT L PP +P SL AS +P+ R+++ PRR S +T R
Sbjct: 247 RSSLSSRSSTTTTLCPPVTPSSLRASRTPSPPTLSRRSARGRRPRRPSRRPST-RDTRAA 423
Query: 250 RTSVFSSMSSSDSGAGSQTGRTRVDGKEF 278
RT S +S S A S G F
Sbjct: 424 RTGGSSPLSVSSGCAQSYGWNATTAGMRF 510
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.125 0.332
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,008,713
Number of Sequences: 63676
Number of extensions: 146302
Number of successful extensions: 2280
Number of sequences better than 10.0: 247
Number of HSP's better than 10.0 without gapping: 2013
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2189
length of query: 339
length of database: 12,639,632
effective HSP length: 98
effective length of query: 241
effective length of database: 6,399,384
effective search space: 1542251544
effective search space used: 1542251544
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0238.4


