

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0238.21
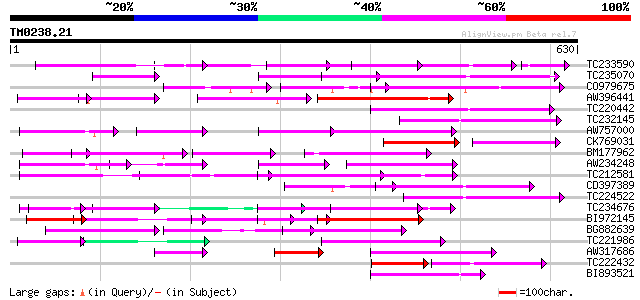
(630 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC233590 similar to UP|Q6K9T2 (Q6K9T2) Pentatricopeptide repeat-... 179 5e-46
TC235070 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 162 3e-40
CO979675 148 6e-36
AW396441 148 8e-36
TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 143 2e-34
TC232145 123 3e-28
AW757000 similar to PIR|C86440|C86 PPR-repeat protein [imported]... 117 1e-26
CK769031 75 4e-26
BM177962 113 2e-25
AW234248 108 5e-24
TC212581 weakly similar to UP|Q6K957 (Q6K957) Pentatricopeptide ... 108 5e-24
CD397389 similar to GP|6691205|gb| F1K23.11 {Arabidopsis thalian... 105 6e-23
TC224522 weakly similar to UP|Q9LUL5 (Q9LUL5) Emb|CAB66100.1, pa... 105 6e-23
TC234676 104 1e-22
BI972145 weakly similar to GP|15408849|db P0672D08.11 {Oryza sat... 103 2e-22
BG882639 102 4e-22
TC221986 weakly similar to GB|BAC16401.1|23237826|AP003749 selen... 102 7e-22
AW317686 101 9e-22
TC222432 62 1e-21
BI893521 100 3e-21
>TC233590 similar to UP|Q6K9T2 (Q6K9T2) Pentatricopeptide repeat-containing
protein-like, partial (5%)
Length = 1094
Score = 179 bits (454), Expect(2) = 5e-46
Identities = 98/280 (35%), Positives = 149/280 (53%), Gaps = 2/280 (0%)
Frame = +2
Query: 286 GDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVL 345
GD + F Q P + SWTS+I GY RNG G AL +F M ++ ++ D L
Sbjct: 23 GDMESGAELFSQMPKSDSCSWTSLIRGYARNGMGYEALGVFKQMIKHQVRPDQFTLSTCL 202
Query: 346 HACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEK-DL 404
ACA++A L HG+ +H+ ++ + V ++VNMY+KCG LE + F I K D+
Sbjct: 203 FACATIASLKHGRQIHAFLVLNNIKPNTIVVCAIVNMYSKCGSLETARRVFNFIGNKQDV 382
Query: 405 VSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAFFRS 464
V WN+M+ A +G EA+ + M+ GVKP++ TF G+L C H GL+ EG F+S
Sbjct: 383 VLWNTMILALAHYGYGIEAIMMLYNMLKIGVKPNKGTFVGILNACCHSGLVQEGLQLFKS 562
Query: 465 MSSEFGLSHGMDHVACMVDMLGRGGYVAEA-QSLAKKYSKTSGARTNSYEVLLGACHAHG 523
M+SE G+ +H + ++LG+ E+ + L K NS +G C HG
Sbjct: 563 MTSEHGVVPDQEHYTRLANLLGQARCFNESVKDLQMMDCKPGDHVCNS---SIGVCRMHG 733
Query: 524 DLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAE 563
++ G+ V +L L+P+ Y +LS Y A G+W+ E
Sbjct: 734 NIDHGAEVAAFLIKLQPQSSAAYELLSRTYAALGKWELVE 853
Score = 82.8 bits (203), Expect = 4e-16
Identities = 54/192 (28%), Positives = 91/192 (47%), Gaps = 1/192 (0%)
Frame = +2
Query: 29 KLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAALSACAGGS 88
+LF +MP DS +W ++I Y+ G+ ++L +F M +PD F+ S L ACA +
Sbjct: 44 ELFSQMPKSDSCSWTSLIRGYARNGMGYEALGVFKQMIKHQVRPDQFTLSTCLFACATIA 223
Query: 89 HHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLF 148
G IHA +V++ + + V ++++MY KC AR+VF+ + + +V
Sbjct: 224 SLKHGRQIHAFLVLNNIKPNTIVVCAIVNMYSKCGSLETARRVFNFIGNKQDV------- 382
Query: 149 AYANSSLFGMALEVFRSMPERVEIAWNTMIAGHARRG-EVEACLGLFKEMCESLYQPDQW 207
+ WNTMI A G +EA + L+ M + +P++
Sbjct: 383 -----------------------VLWNTMILALAHYGYGIEAIMMLY-NMLKIGVKPNKG 490
Query: 208 TFSALMNACTES 219
TF ++NAC S
Sbjct: 491 TFVGILNACCHS 526
Score = 72.8 bits (177), Expect = 4e-13
Identities = 46/197 (23%), Positives = 84/197 (42%)
Frame = +2
Query: 161 EVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTESR 220
E+F MP+ +W ++I G+AR G LG+FK+M + +PDQ+T S + AC
Sbjct: 44 ELFSQMPKSDSCSWTSLIRGYARNGMGYEALGVFKQMIKHQVRPDQFTLSTCLFACATIA 223
Query: 221 DMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVSWNAIID 280
+ +G +H F++ + V +I++ Y+K A +FN G
Sbjct: 224 SLKHGRQIHAFLVLNNIKPNTIVVCAIVNMYSKCGSLETARRVFNFIG------------ 367
Query: 281 AHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLV 340
+++V W +MI+ G G A+ M +M + ++ +
Sbjct: 368 ------------------NKQDVVLWNTMILALAHYGYGIEAIMMLYNMLKIGVKPNKGT 493
Query: 341 AGAVLHACASLAILAHG 357
+L+AC ++ G
Sbjct: 494 FVGILNACCHSGLVQEG 544
Score = 52.0 bits (123), Expect = 8e-07
Identities = 24/79 (30%), Positives = 43/79 (54%)
Frame = +2
Query: 380 VNMYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDE 439
V+ YA GD+E A F + + D SW S++ + +G EA+ +F++M+ V+PD+
Sbjct: 2 VSGYAVWGDMESGAELFSQMPKSDSCSWTSLIRGYARNGMGYEALGVFKQMIKHQVRPDQ 181
Query: 440 VTFTGMLMTCSHLGLIDEG 458
T + L C+ + + G
Sbjct: 182 FTLSTCLFACATIASLKHG 238
Score = 24.3 bits (51), Expect(2) = 5e-46
Identities = 18/54 (33%), Positives = 23/54 (42%)
Frame = +1
Query: 569 MLDQGVKKVPGSSWIEIRNVVTAFVSGNNSSPYMADISNILYFLEIEMRHTRPI 622
M D+G K SWIEI N V F + S P + L+ L M P+
Sbjct: 874 MRDRGGKG-QAISWIEIDNKVHTFTVLDASHPLKETTYSALWALSYRMEGHAPL 1032
>TC235070 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (22%)
Length = 875
Score = 162 bits (411), Expect = 3e-40
Identities = 84/265 (31%), Positives = 145/265 (54%), Gaps = 1/265 (0%)
Frame = +2
Query: 347 ACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVS 406
AC+ L G+++H+ +I+ ++VG +LV+ Y+KCG L + +F I ++ +
Sbjct: 2 ACSCLCSFRQGQLLHAHLIKTPFQVNVYVGTALVDFYSKCGHLAEAQRSFISIFSPNVAA 181
Query: 407 WNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAFFRSMS 466
W +++ + HG +EA+ LFR M+ G+ P+ TF G+L C+H GL+ EG F SM
Sbjct: 182 WTALINGYAYHGLGSEAILLFRSMLHQGIVPNAATFVGVLSACNHAGLVCEGLRIFHSMQ 361
Query: 467 SEFGLSHGMDHVACMVDMLGRGGYVAEAQS-LAKKYSKTSGARTNSYEVLLGACHAHGDL 525
+G++ ++H C+VD+LGR G++ EA+ + K + G + LL A D+
Sbjct: 362 RCYGVTPTIEHYTCVVDLLGRSGHLKEAEEFIIKMPIEADGI---IWGALLNASWFWKDM 532
Query: 526 GTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGVKKVPGSSWIEI 585
G E L +L+P +V+LSN+Y G+W + +RK + ++K PG SWIE+
Sbjct: 533 EVGERAAEKLFSLDPNPIFAFVVLSNMYAILGRWGQKTKLRKRLQSLELRKDPGCSWIEL 712
Query: 586 RNVVTAFVSGNNSSPYMADISNILY 610
N + F + + Y S+++Y
Sbjct: 713 NNKIHLFSVEDKTHLY----SDVIY 775
Score = 47.0 bits (110), Expect = 3e-05
Identities = 35/139 (25%), Positives = 62/139 (44%), Gaps = 2/139 (1%)
Frame = +2
Query: 277 AIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQL 336
A++D + K G +A +F N+ +WT++I GY +G G A+ +F M I
Sbjct: 95 ALVDFYSKCGHLAEAQRSFISIFSPNVAAWTALINGYAYHGLGSEAILLFRSMLHQGIVP 274
Query: 337 DNLVAGAVLHACASLAILAHG-KMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDL-EGSAL 394
+ VL AC ++ G ++ HS G+ + +V++ + G L E
Sbjct: 275 NAATFVGVLSACNHAGLVCEGLRIFHSMQRCYGVTPTIEHYTCVVDLLGRSGHLKEAEEF 454
Query: 395 AFCGILEKDLVSWNSMLFA 413
+E D + W ++L A
Sbjct: 455 IIKMPIEADGIIWGALLNA 511
Score = 45.4 bits (106), Expect = 7e-05
Identities = 22/74 (29%), Positives = 41/74 (54%)
Frame = +2
Query: 93 GSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYAN 152
G ++HA ++ + ++ ++ V +L+D Y KC +A++ F + N W +L+ YA
Sbjct: 32 GQLLHAHLIKTPFQVNVYVGTALVDFYSKCGHLAEAQRSFISIFSPNVAAWTALINGYAY 211
Query: 153 SSLFGMALEVFRSM 166
L A+ +FRSM
Sbjct: 212 HGLGSEAILLFRSM 253
Score = 39.3 bits (90), Expect = 0.005
Identities = 25/98 (25%), Positives = 47/98 (47%)
Frame = +2
Query: 215 ACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVS 274
AC+ G ++H +IK+ + + V +++ FY+K ++A F S + N +
Sbjct: 2 ACSCLCSFRQGQLLHAHLIKTPFQVNVYVGTALVDFYSKCGHLAEAQRSFISIFSPNVAA 181
Query: 275 WNAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVG 312
W A+I+ + G +A L F+ + IV + VG
Sbjct: 182 WTALINGYAYHGLGSEAILLFRSMLHQGIVPNAATFVG 295
>CO979675
Length = 732
Score = 148 bits (374), Expect = 6e-36
Identities = 77/218 (35%), Positives = 119/218 (54%), Gaps = 3/218 (1%)
Frame = -2
Query: 402 KDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAF 461
+D+V WN+M+ +HG A+ +F EM +G+KPD++TF + CS+ G+ EG
Sbjct: 731 RDIVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACSYSGMAHEGLQL 552
Query: 462 FRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTS---GARTNSYEVLLGA 518
MSS + + +H C+VD+L R G EA + ++ + TS T ++ L A
Sbjct: 551 LDKMSSLYEIEPKSEHYGCLVDLLSRAGLFGEAMVMIRRITSTSWNGSEETLAWRAFLSA 372
Query: 519 CHAHGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGVKKVP 578
C HG + L LE V YV+LSNLY ASG+ +A VR M ++GV K P
Sbjct: 371 CCNHGQAQLAERAAKRLLRLENHSGV-YVLLSNLYAASGKHSDARRVRNMMRNKGVDKAP 195
Query: 579 GSSWIEIRNVVTAFVSGNNSSPYMADISNILYFLEIEM 616
G S +EI VV+ F++G + P M +I ++L L +++
Sbjct: 194 GCSSVEIDGVVSEFIAGEETHPQMEEIHSVLEILHMQL 81
Score = 44.7 bits (104), Expect = 1e-04
Identities = 35/133 (26%), Positives = 57/133 (42%), Gaps = 13/133 (9%)
Frame = -2
Query: 172 IAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTESRDMLYGCMVH-G 230
+ WN MI+G A G+ + L +F EM ++ +PD TF A+ AC+ Y M H G
Sbjct: 722 VCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACS------YSGMAHEG 561
Query: 231 FVIKSGWSSAMEVK------NSILSFYAKLECPSDAMEMFNSF------GAFNQVSWNAI 278
+ SS E++ ++ ++ +AM M G+ ++W A
Sbjct: 560 LQLLDKMSSLYEIEPKSEHYGCLVDLLSRAGLFGEAMVMIRRITSTSWNGSEETLAWRAF 381
Query: 279 IDAHMKLGDTQKA 291
+ A G Q A
Sbjct: 380 LSACCNHGQAQLA 342
Score = 43.1 bits (100), Expect = 4e-04
Identities = 37/135 (27%), Positives = 62/135 (45%), Gaps = 12/135 (8%)
Frame = -2
Query: 301 KNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHACASLAILAHG--- 357
++IV W +MI G +G+G AL MF +M + I+ D++ AV AC+ + G
Sbjct: 731 RDIVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACSYSGMAHEGLQL 552
Query: 358 --KMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGIL-------EKDLVSWN 408
KM I + Y LV++ ++ G L G A+ + ++ ++W
Sbjct: 551 LDKMSSLYEIEPKSEHY----GCLVDLLSRAG-LFGEAMVMIRRITSTSWNGSEETLAWR 387
Query: 409 SMLFAFGLHGRANEA 423
+ L A HG+A A
Sbjct: 386 AFLSACCNHGQAQLA 342
Score = 41.2 bits (95), Expect = 0.001
Identities = 21/57 (36%), Positives = 31/57 (53%), Gaps = 2/57 (3%)
Frame = -2
Query: 37 RDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAALSAC--AGGSHHG 91
RD V WNAMI+ + G +L +F M + KPD ++ A +AC +G +H G
Sbjct: 731 RDIVCWNAMISGLAMHGDGASALKMFSEMEKTGIKPDDITFIAVFTACSYSGMAHEG 561
>AW396441
Length = 459
Score = 148 bits (373), Expect = 8e-36
Identities = 73/151 (48%), Positives = 100/151 (65%)
Frame = +3
Query: 343 AVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEK 402
AV+ A +++A L HG+ H+ +I+ GLD FV NSLV+MYAKCG +E S AF ++
Sbjct: 9 AVIAAASNIASLRHGQQFHNQVIKMGLDDDPFVTNSLVDMYAKCGSIEESHKAFSSTNQR 188
Query: 403 DLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAFF 462
D+ WNSM+ + HG A +A+ +F M+ GVKP+ VTF G+L CSH GL+D GF F
Sbjct: 189 DIACWNSMISTYAQHGDAAKALEVFERMIMEGVKPNYVTFVGLLSACSHAGLLDLGFHHF 368
Query: 463 RSMSSEFGLSHGMDHVACMVDMLGRGGYVAE 493
SM S+FG+ +DH ACMV +LGR G + E
Sbjct: 369 ESM-SKFGIEPXIDHYACMVSLLGRAGKIYE 458
Score = 48.9 bits (115), Expect = 7e-06
Identities = 32/131 (24%), Positives = 60/131 (45%), Gaps = 4/131 (3%)
Frame = +3
Query: 209 FSALMNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFG 268
F+A++ A + + +G H VIK G V NS++ YAK ++ + F+S
Sbjct: 3 FAAVIAAASNIASLRHGQQFHNQVIKMGLDDDPFVTNSLVDMYAKCGSIEESHKAFSSTN 182
Query: 269 AFNQVSWNAIIDAHMKLGDTQKAFLAFQ----QAPDKNIVSWTSMIVGYTRNGNGELALS 324
+ WN++I + + GD KA F+ + N V++ ++ + G +L
Sbjct: 183 QRDIACWNSMISTYAQHGDAAKALEVFERMIMEGVKPNYVTFVGLLSACSHAGLLDLGFH 362
Query: 325 MFLDMTRNSIQ 335
F M++ I+
Sbjct: 363 HFESMSKFGIE 395
Score = 48.1 bits (113), Expect = 1e-05
Identities = 28/88 (31%), Positives = 42/88 (46%), Gaps = 6/88 (6%)
Frame = +3
Query: 9 FQTTSKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRIS 68
F T S + A+ G I + K F RD WN+MI+ Y+ G ++L +F M +
Sbjct: 102 FVTNSLVDMYAKCGSIEESHKAFSSTNQRDIACWNSMISTYAQHGDAAKALEVFERMIME 281
Query: 69 NSKPDSFSYSAALSACAG------GSHH 90
KP+ ++ LSAC+ G HH
Sbjct: 282 GVKPNYVTFVGLLSACSHAGLLDLGFHH 365
Score = 47.4 bits (111), Expect = 2e-05
Identities = 27/90 (30%), Positives = 40/90 (44%)
Frame = +3
Query: 77 YSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMA 136
++A ++A + + G H V+ G V NSL+DMY KC ++ K F
Sbjct: 3 FAAVIAAASNIASLRHGQQFHNQVIKMGLDDDPFVTNSLVDMYAKCGSIEESHKAFSSTN 182
Query: 137 DSNEVTWCSLLFAYANSSLFGMALEVFRSM 166
+ W S++ YA ALEVF M
Sbjct: 183 QRDIACWNSMISTYAQHGDAAKALEVFERM 272
>TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (25%)
Length = 706
Score = 143 bits (361), Expect = 2e-34
Identities = 68/205 (33%), Positives = 115/205 (55%)
Frame = +1
Query: 401 EKDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFA 460
+K + +W +++ +HG+ EA+ F +M +G+ P+ +TFT +L CSH GL +EG +
Sbjct: 1 KKCVCAWTAIIGGLAIHGKGREALDWFTQMQKAGINPNSITFTAILTACSHAGLTEEGKS 180
Query: 461 FFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTSGARTNSYEVLLGACH 520
F SMSS + + M+H CMVD++GR G + EA+ + A + LL AC
Sbjct: 181 LFESMSSVYNIKPSMEHYGCMVDLMGRAGLLKEAREFIESMPVKPNAAI--WGALLNACQ 354
Query: 521 AHGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGVKKVPGS 580
H G +G+ L L+P+ Y+ L+++Y A+G+W + VR ++ +G+ PG
Sbjct: 355 LHKHFELGKEIGKILIELDPDHSGRYIHLASIYAAAGEWNQVVRVRSQIKHRGLLNHPGC 534
Query: 581 SWIEIRNVVTAFVSGNNSSPYMADI 605
S I + VV F +G+ S P++ +I
Sbjct: 535 SSITLNGVVHEFFAGDGSHPHIQEI 609
Score = 33.1 bits (74), Expect = 0.38
Identities = 25/71 (35%), Positives = 37/71 (51%), Gaps = 5/71 (7%)
Frame = +1
Query: 12 TSKIVSLARSGRICHARKLFDEMP----DRDSVAWNAMITAYSHLGLYQQSLSLFGSM-R 66
T+ I LA G+ A F +M + +S+ + A++TA SH GL ++ SLF SM
Sbjct: 22 TAIIGGLAIHGKGREALDWFTQMQKAGINPNSITFTAILTACSHAGLTEEGKSLFESMSS 201
Query: 67 ISNSKPDSFSY 77
+ N KP Y
Sbjct: 202VYNIKPSMEHY 234
>TC232145
Length = 817
Score = 123 bits (308), Expect = 3e-28
Identities = 68/180 (37%), Positives = 101/180 (55%)
Frame = +3
Query: 434 GVKPDEVTFTGMLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAE 493
GV P+ +TF +L CSH G ID+GF +F SM++ + G++H ACMVD+LGR G + E
Sbjct: 6 GVVPEYITFVSVLSACSHTGKIDDGFKYFNSMANVHNIKPGLEHYACMVDLLGRVGRLEE 185
Query: 494 AQSLAKKYSKTSGARTNSYEVLLGACHAHGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLY 553
A + S + + LLGAC H ++ G V E L LEP+ Y++LSN+Y
Sbjct: 186 ACRFIE--SMPFEPDSLVWGALLGACGKHANVEMGREVAERLFKLEPDNPGNYMLLSNIY 359
Query: 554 CASGQWKEAEIVRKEMLDQGVKKVPGSSWIEIRNVVTAFVSGNNSSPYMADISNILYFLE 613
G +EA+ VR+ M GV+K G SWI+++N F + + S +I +L L+
Sbjct: 360 IRHGMLEEADEVRRLMGINGVRKESGCSWIDVKNRTFVFNANDRSHSRTQEIYGMLQKLK 539
>AW757000 similar to PIR|C86440|C86 PPR-repeat protein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 442
Score = 117 bits (294), Expect = 1e-26
Identities = 54/140 (38%), Positives = 83/140 (58%)
Frame = +3
Query: 357 GKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGL 416
GK +H+ I + VG +L+ MYAKCG +E S F G+ EKD SW S++ +
Sbjct: 9 GKWIHNYIDENRIKVDAVVGTALIEMYAKCGCIEKSFEIFNGLKEKDTTSWTSIICGLAM 188
Query: 417 HGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMD 476
+G+ +EA+ LF+ M G+KPD++TF +L CSH GL++EG F SMSS + + ++
Sbjct: 189 NGKPSEALELFKAMQTCGLKPDDITFVAVLSACSHAGLVEEGRKLFHSMSSMYHIEPNLE 368
Query: 477 HVACMVDMLGRGGYVAEAQS 496
H C +D+LG + +S
Sbjct: 369 HYGCFIDLLGEPDFYKRQRS 428
Score = 54.3 bits (129), Expect = 2e-07
Identities = 30/87 (34%), Positives = 47/87 (53%), Gaps = 1/87 (1%)
Frame = +3
Query: 277 AIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQL 336
A+I+ + K G +K+F F +K+ SWTS+I G NG AL +F M ++
Sbjct: 72 ALIEMYAKCGCIEKSFEIFNGLKEKDTTSWTSIICGLAMNGKPSEALELFKAMQTCGLKP 251
Query: 337 DNLVAGAVLHACASLAILAHG-KMVHS 362
D++ AVL AC+ ++ G K+ HS
Sbjct: 252 DDITFVAVLSACSHAGLVEEGRKLFHS 332
Score = 47.0 bits (110), Expect = 3e-05
Identities = 24/79 (30%), Positives = 40/79 (50%)
Frame = +3
Query: 141 VTWCSLLFAYANSSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCES 200
V +L+ YA + E+F + E+ +W ++I G A G+ L LFK M
Sbjct: 60 VVGTALIEMYAKCGCIEKSFEIFNGLKEKDTTSWTSIICGLAMNGKPSEALELFKAMQTC 239
Query: 201 LYQPDQWTFSALMNACTES 219
+PD TF A+++AC+ +
Sbjct: 240 GLKPDDITFVAVLSACSHA 296
Score = 46.6 bits (109), Expect = 3e-05
Identities = 30/114 (26%), Positives = 57/114 (49%), Gaps = 4/114 (3%)
Frame = +3
Query: 12 TSKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSK 71
T+ I A+ G I + ++F+ + ++D+ +W ++I + G ++L LF +M+ K
Sbjct: 69 TALIEMYAKCGCIEKSFEIFNGLKEKDTTSWTSIICGLAMNGKPSEALELFKAMQTCGLK 248
Query: 72 PDSFSYSAALSACAGGSHHGF----GSVIHALVVVSGYRSSLPVANSLIDMYGK 121
PD ++ A LSAC SH G + H++ + +L ID+ G+
Sbjct: 249 PDDITFVAVLSAC---SHAGLVEEGRKLFHSMSSMYHIEPNLEHYGCFIDLLGE 401
Score = 38.5 bits (88), Expect = 0.009
Identities = 28/114 (24%), Positives = 55/114 (47%), Gaps = 4/114 (3%)
Frame = +3
Query: 93 GSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYAN 152
G IH + + + V +LI+MY KC + ++F+ + + + +W S++ A
Sbjct: 9 GKWIHNYIDENRIKVDAVVGTALIEMYAKCGCIEKSFEIFNGLKEKDTTSWTSIICGLAM 188
Query: 153 SSLFGMALEVFRSMP----ERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLY 202
+ ALE+F++M + +I + +++ + G VE LF M S+Y
Sbjct: 189 NGKPSEALELFKAMQTCGLKPDDITFVAVLSACSHAGLVEEGRKLFHSM-SSMY 347
>CK769031
Length = 827
Score = 75.1 bits (183), Expect(2) = 4e-26
Identities = 37/85 (43%), Positives = 52/85 (60%), Gaps = 1/85 (1%)
Frame = +2
Query: 416 LHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAFFRSMSS-EFGLSHG 474
++G EA+ F ++V SG +P+EVTF + C H GL+DEG +F M+S + LS
Sbjct: 23 INGYGKEALKRFEDLVESGARPNEVTFLAVYTACCHNGLVDEGRKYFNEMTSPHYNLSPC 202
Query: 475 MDHVACMVDMLGRGGYVAEAQSLAK 499
++H CMVD+L R G V EA L K
Sbjct: 203 LEHYGCMVDLLCRAGLVGEAVELIK 277
Score = 62.0 bits (149), Expect(2) = 4e-26
Identities = 34/98 (34%), Positives = 54/98 (54%)
Frame = +1
Query: 515 LLGACHAHGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGV 574
LL AC+ G++G + + L+ +E + YV+LSNLY + +W E +R+ M +G+
Sbjct: 298 LLSACNT*GNVGFTQEMLKSLQNVEFQDSGIYVLLSNLYATNKKWAEVRSIRRLMKQKGI 477
Query: 575 KKVPGSSWIEIRNVVTAFVSGNNSSPYMADISNILYFL 612
K PGSS I + + F+ G+ S P DI +L L
Sbjct: 478 SKAPGSSIISVDGMSHEFLVGDISHPQSEDIYILLNIL 591
>BM177962
Length = 421
Score = 113 bits (283), Expect = 2e-25
Identities = 57/141 (40%), Positives = 84/141 (59%)
Frame = +2
Query: 328 DMTRNSIQLDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCG 387
D NS+ + N VL ACA LA L GK++H I+RRGLD L V N+L+ MY +CG
Sbjct: 14 DSVPNSVTMVN-----VLQACAGLAALEQGKLIHGYILRRGLDSILPVLNALITMYGRCG 178
Query: 388 DLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLM 447
++ F + +D+VSWNS++ +G+HG +A+ +F M+ G P ++F +L
Sbjct: 179 EILMGQRVFDNMKNRDVVSWNSLISIYGMHGFGKKAIQIF*NMIHQGGSPSYISFITVLG 358
Query: 448 TCSHLGLIDEGFAFFRSMSSE 468
CSH GL++EG F SM S+
Sbjct: 359 ACSHAGLVEEGKILFESMLSK 421
Score = 64.7 bits (156), Expect = 1e-10
Identities = 43/133 (32%), Positives = 70/133 (52%), Gaps = 4/133 (3%)
Frame = +2
Query: 69 NSKPDSFSYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDA 128
+S P+S + L ACAG + G +IH ++ G S LPV N+LI MYG+C +
Sbjct: 14 DSVPNSVTMVNVLQACAGLAALEQGKLIHGYILRRGLDSILPVLNALITMYGRCGEILMG 193
Query: 129 RKVFDEMADSNEVTWCSLLFAYANSSLFGMALEVFRSMPER----VEIAWNTMIAGHARR 184
++VFD M + + V+W SL+ Y A+++F +M + I++ T++ +
Sbjct: 194 QRVFDNMKNRDVVSWNSLISIYGMHGFGKKAIQIF*NMIHQGGSPSYISFITVLGACSHA 373
Query: 185 GEVEACLGLFKEM 197
G VE LF+ M
Sbjct: 374 GLVEEGKILFESM 412
Score = 48.9 bits (115), Expect = 7e-06
Identities = 25/77 (32%), Positives = 40/77 (51%)
Frame = +2
Query: 15 IVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDS 74
I R G I +++FD M +RD V+WN++I+ Y G ++++ +F +M P
Sbjct: 155 ITMYGRCGEILMGQRVFDNMKNRDVVSWNSLISIYGMHGFGKKAIQIF*NMIHQGGSPSY 334
Query: 75 FSYSAALSACAGGSHHG 91
S+ L AC SH G
Sbjct: 335 ISFITVLGAC---SHAG 376
Score = 43.5 bits (101), Expect = 3e-04
Identities = 22/92 (23%), Positives = 46/92 (49%)
Frame = +2
Query: 204 PDQWTFSALMNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEM 263
P+ T ++ AC + G ++HG++++ G S + V N++++ Y + +
Sbjct: 23 PNSVTMVNVLQACAGLAALEQGKLIHGYILRRGLDSILPVLNALITMYGRCGEILMGQRV 202
Query: 264 FNSFGAFNQVSWNAIIDAHMKLGDTQKAFLAF 295
F++ + VSWN++I + G +KA F
Sbjct: 203 FDNMKNRDVVSWNSLISIYGMHGFGKKAIQIF 298
>AW234248
Length = 399
Score = 108 bits (271), Expect = 5e-24
Identities = 52/123 (42%), Positives = 73/123 (59%)
Frame = +2
Query: 375 VGNSLVNMYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMVASG 434
V SLV+MYAKCG++E + F + SWN+M+ HG A EA+ F EM + G
Sbjct: 2 VMTSLVDMYAKCGNIEDARGLFKRTNTSRIASWNAMIVGLAQHGNAEEALQFFEEMKSRG 181
Query: 435 VKPDEVTFTGMLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEA 494
V PD VTF G+L CSH GL+ E + F SM +G+ ++H +C+VD L R G + EA
Sbjct: 182 VTPDRVTFIGVLSACSHSGLVSEAYENFYSMQKIYGIEPEIEHYSCLVDALSRAGRIREA 361
Query: 495 QSL 497
+ +
Sbjct: 362 EKV 370
Score = 49.3 bits (116), Expect = 5e-06
Identities = 24/79 (30%), Positives = 42/79 (52%)
Frame = +2
Query: 277 AIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQL 336
+++D + K G+ + A F++ I SW +MIVG ++GN E AL F +M +
Sbjct: 11 SLVDMYAKCGNIEDARGLFKRTNTSRIASWNAMIVGLAQHGNAEEALQFFEEMKSRGVTP 190
Query: 337 DNLVAGAVLHACASLAILA 355
D + VL AC+ +++
Sbjct: 191 DRVTFIGVLSACSHSGLVS 247
Score = 49.3 bits (116), Expect = 5e-06
Identities = 35/128 (27%), Positives = 55/128 (42%), Gaps = 4/128 (3%)
Frame = +2
Query: 12 TSKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSK 71
TS + A+ G I AR LF +WNAMI + G +++L F M+
Sbjct: 8 TSLVDMYAKCGNIEDARGLFKRTNTSRIASWNAMIVGLAQHGNAEEALQFFEEMKSRGVT 187
Query: 72 PDSFSYSAALSACAGGSHHGFGSV----IHALVVVSGYRSSLPVANSLIDMYGKCLKPHD 127
PD ++ LSAC SH G S +++ + G + + L+D + + +
Sbjct: 188 PDRVTFIGVLSAC---SHSGLVSEAYENFYSMQKIYGIEPEIEHYSCLVDALSRAGRIRE 358
Query: 128 ARKVFDEM 135
A KV M
Sbjct: 359 AEKVISSM 382
Score = 47.4 bits (111), Expect = 2e-05
Identities = 33/109 (30%), Positives = 45/109 (41%)
Frame = +2
Query: 111 VANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYANSSLFGMALEVFRSMPERV 170
V SL+DMY KC DAR +F R+ R+
Sbjct: 2 VMTSLVDMYAKCGNIEDARGLFK------------------------------RTNTSRI 91
Query: 171 EIAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTES 219
+WN MI G A+ G E L F+EM PD+ TF +++AC+ S
Sbjct: 92 A-SWNAMIVGLAQHGNAEEALQFFEEMKSRGVTPDRVTFIGVLSACSHS 235
>TC212581 weakly similar to UP|Q6K957 (Q6K957) Pentatricopeptide (PPR)
repeat-containing protein-like, partial (8%)
Length = 1044
Score = 108 bits (271), Expect = 5e-24
Identities = 68/224 (30%), Positives = 114/224 (50%), Gaps = 2/224 (0%)
Frame = +3
Query: 276 NAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQ 335
N ++ + K G + A L F Q P +N+ +WT++I + R G+ A MF + + +
Sbjct: 222 NNLLSLYAKFGHFRHAHLLFDQMPQRNVFTWTTLISSHFRTGSLPKAFEMFNHICALNER 401
Query: 336 LDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSAL- 394
+ +L ACA+ ++ G +H ++R GL++ F G+S+V MY G G A
Sbjct: 402 PNEYTFSVLLRACATPSLWNVGLQIHGLLVRSGLERNKFSGSSIVYMYFNSGSNLGDACC 581
Query: 395 AFCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREM-VASGVKPDEVTFTGMLMTCSHLG 453
AF +LE+DLV+WN M+ F G + LF EM G+KPD+ TF +L CS L
Sbjct: 582 AFHDLLERDLVAWNVMISGFARVGDFSMVHRLFSEMWGVEGLKPDDCTFVSLLKCCSSLK 761
Query: 454 LIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSL 497
+ + ++S+FG + +VD+ + G V+ + +
Sbjct: 762 ELKQ----IHGLASKFGAEVDVVVGNALVDL*NKHGDVSSCRKV 881
Score = 101 bits (251), Expect = 1e-21
Identities = 82/287 (28%), Positives = 129/287 (44%), Gaps = 5/287 (1%)
Frame = +3
Query: 12 TSKIVSL-ARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNS 70
T+ ++SL A+ G HA LFD+MP R+ W +I+++ G ++ +F + N
Sbjct: 219 TNNLLSLYAKFGHFRHAHLLFDQMPQRNVFTWTTLISSHFRTGSLPKAFEMFNHICALNE 398
Query: 71 KPDSFSYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARK 130
+P+ +++S L ACA S G IH L+V SG
Sbjct: 399 RPNEYTFSVLLRACATPSLWNVGLQIHGLLVRSG-------------------------- 500
Query: 131 VFDEMADSNEVTWCSLLFAYANS-SLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEA 189
+ N+ + S+++ Y NS S G A F + ER +AWN MI+G AR G+
Sbjct: 501 -----LERNKFSGSSIVYMYFNSGSNLGDACCAFHDLLERDLVAWNVMISGFARVGDFSM 665
Query: 190 CLGLFKEM--CESLYQPDQWTFSALMNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSI 247
LF EM E L +PD TF +L+ C+ +++ +HG K G + V N++
Sbjct: 666 VHRLFSEMWGVEGL-KPDDCTFVSLLKCCSSLKELK---QIHGLASKFGAEVDVVVGNAL 833
Query: 248 LSFYAKLECPSDAMEMFNSFGAFNQVSWNAIIDAH-MKLGDTQKAFL 293
+ K S ++F+S W+ II + M G AFL
Sbjct: 834 VDL*NKHGDVSSCRKVFDSKKEKYNFVWSLIISGYSMNKG*GSCAFL 974
Score = 95.1 bits (235), Expect = 8e-20
Identities = 72/274 (26%), Positives = 121/274 (43%), Gaps = 1/274 (0%)
Frame = +3
Query: 145 SLLFAYANSSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQP 204
+LL YA F A +F MP+R W T+I+ H R G + +F +C +P
Sbjct: 225 NLLSLYAKFGHFRHAHLLFDQMPQRNVFTWTTLISSHFRTGSLPKAFEMFNHICALNERP 404
Query: 205 DQWTFSALMNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEMF 264
+++TFS L+ AC G +HG +++SG +SI+ Y
Sbjct: 405 NEYTFSVLLRACATPSLWNVGLQIHGLLVRSGLERNKFSGSSIVYMY------------- 545
Query: 265 NSFGAFNQVSWNAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALS 324
FN S LGD A AF ++++V+W MI G+ R G+ +
Sbjct: 546 -----FNSGS---------NLGD---ACCAFHDLLERDLVAWNVMISGFARVGDFSMVHR 674
Query: 325 MFLDMTR-NSIQLDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMY 383
+F +M ++ D+ ++L C+SL L K +H + G + + VGN+LV++
Sbjct: 675 LFSEMWGVEGLKPDDCTFVSLLKCCSSLKEL---KQIHGLASKFGAEVDVVVGNALVDL* 845
Query: 384 AKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLH 417
K GD+ F EK W+ ++ + ++
Sbjct: 846 NKHGDVSSCRKVFDSKKEKYNFVWSLIISGYSMN 947
>CD397389 similar to GP|6691205|gb| F1K23.11 {Arabidopsis thaliana}, partial
(16%)
Length = 626
Score = 105 bits (262), Expect = 6e-23
Identities = 58/179 (32%), Positives = 96/179 (53%), Gaps = 2/179 (1%)
Frame = -2
Query: 407 WNSMLFAFGLHGRANEAMCLFREMVAS-GVKPDEVTFTGMLMTCSHLGLIDEGFAFFRSM 465
W SM+ +G +G +EA+ LF +M G+ P+ VT L C+H GL+D+G+ +SM
Sbjct: 625 WTSMMDGYGKNGFPDEALELFVKMPTEYGIVPNYVTLLSALSACAHAGLVDKGWEIIQSM 446
Query: 466 SSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTSGARTNSYEVLLGACHAHGDL 525
+E+ + GM+H ACMVD+LGR G + +A + + ++ + LL +C HG++
Sbjct: 445 ENEYLVKPGMEHYACMVDLLGRAGMLNQAWEFIMRIPEK--PISDVWAALLSSCRLHGNI 272
Query: 526 GTGS-SVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGVKKVPGSSWI 583
+ E K + YV LSN A+G W+ +R+ M ++G+ K SW+
Sbjct: 271 ELAKLAANELFKLNATGRPGAYVALSNTLVAAG*WESVTELREIMKERGISKDTXRSWV 95
Score = 42.0 bits (97), Expect = 8e-04
Identities = 35/132 (26%), Positives = 61/132 (45%), Gaps = 7/132 (5%)
Frame = -2
Query: 306 WTSMIVGYTRNGNGELALSMFLDM-TRNSIQLDNLVAGAVLHACASLAILAHG-----KM 359
WTSM+ GY +NG + AL +F+ M T I + + + L ACA ++ G M
Sbjct: 625 WTSMMDGYGKNGFPDEALELFVKMPTEYGIVPNYVTLLSALSACAHAGLVDKGWEIIQSM 446
Query: 360 VHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVS-WNSMLFAFGLHG 418
+ +++ G++ Y +V++ + G L + I EK + W ++L + LHG
Sbjct: 445 ENEYLVKPGMEHYA----CMVDLLGRAGMLNQAWEFIMRIPEKPISDVWAALLSSCRLHG 278
Query: 419 RANEAMCLFREM 430
A E+
Sbjct: 277 NIELAKLAANEL 242
>TC224522 weakly similar to UP|Q9LUL5 (Q9LUL5) Emb|CAB66100.1, partial (18%)
Length = 534
Score = 105 bits (262), Expect = 6e-23
Identities = 57/179 (31%), Positives = 90/179 (49%)
Frame = +2
Query: 438 DEVTFTGMLMTCSHLGLIDEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSL 497
D +TF +L C G ++E F M +G+ +H AC+VD++ R G + A +
Sbjct: 2 DGITFLSLLSACCRAGKVNESMNLFSLMVDNYGIPPRSEHYACLVDVMSRAGQLQRACKI 181
Query: 498 AKKYSKTSGARTNSYEVLLGACHAHGDLGTGSSVGEYLKTLEPEKEVGYVMLSNLYCASG 557
+ A ++ + +L AC H ++ G + L+P YVMLSN+Y A+G
Sbjct: 182 INEMPFK--ADSSIWGAVLAACSVHLNVELGELAARRILNLDPFNSGAYVMLSNIYAAAG 355
Query: 558 QWKEAEIVRKEMLDQGVKKVPGSSWIEIRNVVTAFVSGNNSSPYMADISNILYFLEIEM 616
+WK+ +R M +QGVKK SW++I N FV G+ S P + DI L + + M
Sbjct: 356 KWKDVHRIRVLMKEQGVKKQTAYSWLQIGNKTHYFVGGDPSHPNINDIHVALRRITLHM 532
>TC234676
Length = 914
Score = 104 bits (259), Expect = 1e-22
Identities = 61/220 (27%), Positives = 107/220 (47%)
Frame = +1
Query: 276 NAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQ 335
++++ + K G + + F +N++SWT+MI Y NG AL + M + +
Sbjct: 73 SSLMTMYSKCGVVEYSRRLFDNMEQRNVISWTAMIDSYIENGYLCEALGVIRSMQLSKHR 252
Query: 336 LDNLVAGAVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALA 395
D++ G +L C ++ GK +H I++R FV L+NMY GD+ + L
Sbjct: 253 PDSVAIGRMLSVCGERKLVKLGKEIHGQILKRDFTSVHFVSAELINMYGFFGDINKANLV 432
Query: 396 FCGILEKDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLI 455
F + K ++W +++ A+G + +A+ LF +M S P+ TF +L C G +
Sbjct: 433 FNAVPVKGSMTWTALIRAYGYNELYQDAVNLFDQMRYS---PNHFTFEAILSICDKAGFV 603
Query: 456 DEGFAFFRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQ 495
D+ F SM + + +H A MV +L G + +AQ
Sbjct: 604 DDACRIFNSM-PRYKIEASKEHFAIMVRLLTHNGQLEKAQ 720
Score = 80.5 bits (197), Expect = 2e-15
Identities = 46/155 (29%), Positives = 80/155 (50%)
Frame = +1
Query: 12 TSKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSK 71
+S + ++ G + ++R+LFD M R+ ++W AMI +Y G ++L + SM++S +
Sbjct: 73 SSLMTMYSKCGVVEYSRRLFDNMEQRNVISWTAMIDSYIENGYLCEALGVIRSMQLSKHR 252
Query: 72 PDSFSYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKV 131
PDS + LS C G IH ++ + S V+ LI+MYG + A V
Sbjct: 253 PDSVAIGRMLSVCGERKLVKLGKEIHGQILKRDFTSVHFVSAELINMYGFFGDINKANLV 432
Query: 132 FDEMADSNEVTWCSLLFAYANSSLFGMALEVFRSM 166
F+ + +TW +L+ AY + L+ A+ +F M
Sbjct: 433 FNAVPVKGSMTWTALIRAYGYNELYQDAVNLFDQM 537
Score = 68.6 bits (166), Expect = 8e-12
Identities = 56/237 (23%), Positives = 95/237 (39%)
Frame = +1
Query: 93 GSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYAN 152
G IHA + + ++ VA+SL+ MY KC +R++FD M N ++W +++ +Y
Sbjct: 13 GMQIHAYALKHWFLPNVSVASSLMTMYSKCGVVEYSRRLFDNMEQRNVISWTAMIDSYIE 192
Query: 153 SSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSAL 212
+ AL V RSM S ++PD +
Sbjct: 193 NGYLCEALGVIRSMQ-------------------------------LSKHRPDSVAIGRM 279
Query: 213 MNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQ 272
++ C E + + G +HG ++K ++S V + E+ N +G F
Sbjct: 280 LSVCGERKLVKLGKEIHGQILKRDFTSVHFV----------------SAELINMYGFF-- 405
Query: 273 VSWNAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDM 329
GD KA L F P K ++WT++I Y N + A+++F M
Sbjct: 406 -------------GDINKANLVFNAVPVKGSMTWTALIRAYGYNELYQDAVNLFDQM 537
Score = 56.6 bits (135), Expect = 3e-08
Identities = 30/102 (29%), Positives = 54/102 (52%)
Frame = +1
Query: 357 GKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGL 416
G +H+ ++ + V +SL+ MY+KCG +E S F + +++++SW +M+ ++
Sbjct: 13 GMQIHAYALKHWFLPNVSVASSLMTMYSKCGVVEYSRRLFDNMEQRNVISWTAMIDSYIE 192
Query: 417 HGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEG 458
+G EA+ + R M S +PD V ML C L+ G
Sbjct: 193 NGYLCEALGVIRSMQLSKHRPDSVAIGRMLSVCGERKLVKLG 318
Score = 48.9 bits (115), Expect = 7e-06
Identities = 25/63 (39%), Positives = 38/63 (59%)
Frame = +1
Query: 22 GRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAAL 81
G I A +F+ +P + S+ W A+I AY + LYQ +++LF MR S P+ F++ A L
Sbjct: 406 GDINKANLVFNAVPVKGSMTWTALIRAYGYNELYQDAVNLFDQMRYS---PNHFTFEAIL 576
Query: 82 SAC 84
S C
Sbjct: 577 SIC 585
>BI972145 weakly similar to GP|15408849|db P0672D08.11 {Oryza sativa
(japonica cultivar-group)}, partial (7%)
Length = 390
Score = 103 bits (257), Expect = 2e-22
Identities = 47/117 (40%), Positives = 79/117 (67%)
Frame = +3
Query: 343 AVLHACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEK 402
+++ AC +++ L HGK +H +RRG+ ++VG++L++MYAKCG ++ + F +
Sbjct: 33 SLIPACGNISALMHGKEIHCFSLRRGIFDDVYVGSALIDMYAKCGRIQLARRCFDKMSAL 212
Query: 403 DLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGF 459
+LVSWN+++ + +HG+A E M +F M+ SG KPD VTFT +L C+ GL +EG+
Sbjct: 213 NLVSWNAVMKGYAMHGKAKETMEMFHMMLQSGQKPDLVTFTCVLSACAQNGLTEEGW 383
Score = 57.8 bits (138), Expect = 1e-08
Identities = 37/149 (24%), Positives = 67/149 (44%)
Frame = +3
Query: 71 KPDSFSYSAALSACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARK 130
+P++ + + + AC S G IH + G + V ++LIDMY KC + AR+
Sbjct: 9 EPNAVTIPSLIPACGNISALMHGKEIHCFSLRRGIFDDVYVGSALIDMYAKCGRIQLARR 188
Query: 131 VFDEMADSNEVTWCSLLFAYANSSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEAC 190
FD+M+ N V+W N ++ G+A G+ +
Sbjct: 189 CFDKMSALNLVSW-------------------------------NAVMKGYAMHGKAKET 275
Query: 191 LGLFKEMCESLYQPDQWTFSALMNACTES 219
+ +F M +S +PD TF+ +++AC ++
Sbjct: 276 MEMFHMMLQSGQKPDLVTFTCVLSACAQN 362
Score = 53.9 bits (128), Expect = 2e-07
Identities = 26/67 (38%), Positives = 42/67 (61%)
Frame = +3
Query: 19 ARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYS 78
A+ GRI AR+ FD+M + V+WNA++ Y+ G ++++ +F M S KPD +++
Sbjct: 156 AKCGRIQLARRCFDKMSALNLVSWNAVMKGYAMHGKAKETMEMFHMMLQSGQKPDLVTFT 335
Query: 79 AALSACA 85
LSACA
Sbjct: 336 CVLSACA 356
Score = 51.6 bits (122), Expect = 1e-06
Identities = 31/119 (26%), Positives = 59/119 (49%), Gaps = 4/119 (3%)
Frame = +3
Query: 203 QPDQWTFSALMNACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAME 262
+P+ T +L+ AC +++G +H F ++ G + V ++++ YAK A
Sbjct: 9 EPNAVTIPSLIPACGNISALMHGKEIHCFSLRRGIFDDVYVGSALIDMYAKCGRIQLARR 188
Query: 263 MFNSFGAFNQVSWNAIIDA---HMKLGDTQKAF-LAFQQAPDKNIVSWTSMIVGYTRNG 317
F+ A N VSWNA++ H K +T + F + Q ++V++T ++ +NG
Sbjct: 189 CFDKMSALNLVSWNAVMKGYAMHGKAKETMEMFHMMLQSGQKPDLVTFTCVLSACAQNG 365
Score = 45.8 bits (107), Expect = 6e-05
Identities = 25/82 (30%), Positives = 41/82 (49%)
Frame = +3
Query: 276 NAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQ 335
+A+ID + K G Q A F + N+VSW +++ GY +G + + MF M ++ +
Sbjct: 135 SALIDMYAKCGRIQLARRCFDKMSALNLVSWNAVMKGYAMHGKAKETMEMFHMMLQSGQK 314
Query: 336 LDNLVAGAVLHACASLAILAHG 357
D + VL ACA + G
Sbjct: 315 PDLVTFTCVLSACAQNGLTEEG 380
>BG882639
Length = 414
Score = 102 bits (255), Expect = 4e-22
Identities = 57/138 (41%), Positives = 81/138 (58%)
Frame = +1
Query: 304 VSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHACASLAILAHGKMVHSC 363
VSWT+MI GY +N AL++FL M + ++ +VL ACA + L G VH C
Sbjct: 1 VSWTAMISGYVQNKRFMDALNLFLLMFNSGTCPNHFTFSSVLDACAGCSSLLTGMQVHLC 180
Query: 364 IIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVSWNSMLFAFGLHGRANEA 423
+I+ G+ + + SLV+MYAKCGD++ + F I K+LVSWNS++ +G A A
Sbjct: 181 VIKSGIPEDVISLTSLVDMYAKCGDMDAAFRVFESIPNKNLVSWNSIIGGCARNGIATRA 360
Query: 424 MCLFREMVASGVKPDEVT 441
+ F M +GV PDEVT
Sbjct: 361 LEEFDRMKKAGVTPDEVT 414
Score = 78.6 bits (192), Expect = 8e-15
Identities = 51/168 (30%), Positives = 75/168 (44%)
Frame = +1
Query: 172 IAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTESRDMLYGCMVHGF 231
++W MI+G+ + L LF M S P+ +TFS++++AC +L G VH
Sbjct: 1 VSWTAMISGYVQNKRFMDALNLFLLMFNSGTCPNHFTFSSVLDACAGCSSLLTGMQVHLC 180
Query: 232 VIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVSWNAIIDAHMKLGDTQKA 291
VIKSG P D +S +++D + K GD A
Sbjct: 181 VIKSG-------------------IPEDV------------ISLTSLVDMYAKCGDMDAA 267
Query: 292 FLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNL 339
F F+ P+KN+VSW S+I G RNG AL F M + + D +
Sbjct: 268 FRVFESIPNKNLVSWNSIIGGCARNGIATRALEEFDRMKKAGVTPDEV 411
Score = 77.8 bits (190), Expect = 1e-14
Identities = 45/127 (35%), Positives = 68/127 (53%)
Frame = +1
Query: 40 VAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAALSACAGGSHHGFGSVIHAL 99
V+W AMI+ Y + +L+LF M S + P+ F++S+ L ACAG S G +H
Sbjct: 1 VSWTAMISGYVQNKRFMDALNLFLLMFNSGTCPNHFTFSSVLDACAGCSSLLTGMQVHLC 180
Query: 100 VVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVTWCSLLFAYANSSLFGMA 159
V+ SG + SL+DMY KC A +VF+ + + N V+W S++ A + + A
Sbjct: 181 VIKSGIPEDVISLTSLVDMYAKCGDMDAAFRVFESIPNKNLVSWNSIIGGCARNGIATRA 360
Query: 160 LEVFRSM 166
LE F M
Sbjct: 361 LEEFDRM 381
Score = 35.8 bits (81), Expect = 0.059
Identities = 17/62 (27%), Positives = 36/62 (57%)
Frame = +1
Query: 12 TSKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSK 71
TS + A+ G + A ++F+ +P+++ V+WN++I + G+ ++L F M+ +
Sbjct: 220 TSLVDMYAKCGDMDAAFRVFESIPNKNLVSWNSIIGGCARNGIATRALEEFDRMKKAGVT 399
Query: 72 PD 73
PD
Sbjct: 400 PD 405
>TC221986 weakly similar to GB|BAC16401.1|23237826|AP003749 selenium-binding
protein-like {Oryza sativa (japonica cultivar-group);} ,
partial (10%)
Length = 853
Score = 102 bits (253), Expect = 7e-22
Identities = 43/138 (31%), Positives = 82/138 (59%)
Frame = +1
Query: 347 ACASLAILAHGKMVHSCIIRRGLDKYLFVGNSLVNMYAKCGDLEGSALAFCGILEKDLVS 406
AC + L G+ VH ++ G + F+G +L++MY+KCG+L+ + F + + L +
Sbjct: 4 ACTEMGSLKLGRRVHDFALKNGFELEPFLGTALIDMYSKCGNLDDARTVFDMMQMRTLAT 183
Query: 407 WNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAFFRSMS 466
WN+M+ + G+HG +EA+ +F EM + PD +TF G+L C ++ ++ +F M+
Sbjct: 184 WNTMITSLGVHGYRDEALSIFEEMEKANEVPDAITFVGVLSACVYMNDLELAQKYFNLMT 363
Query: 467 SEFGLSHGMDHVACMVDM 484
+G++ ++H CMV++
Sbjct: 364 DHYGITPILEHYTCMVEI 417
Score = 51.6 bits (122), Expect = 1e-06
Identities = 27/76 (35%), Positives = 40/76 (52%)
Frame = +1
Query: 9 FQTTSKIVSLARSGRICHARKLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRIS 68
F T+ I ++ G + AR +FD M R WN MIT+ G ++LS+F M +
Sbjct: 85 FLGTALIDMYSKCGNLDDARTVFDMMQMRTLATWNTMITSLGVHGYRDEALSIFEEMEKA 264
Query: 69 NSKPDSFSYSAALSAC 84
N PD+ ++ LSAC
Sbjct: 265 NEVPDAITFVGVLSAC 312
Score = 49.3 bits (116), Expect = 5e-06
Identities = 37/140 (26%), Positives = 52/140 (36%)
Frame = +1
Query: 83 ACAGGSHHGFGSVIHALVVVSGYRSSLPVANSLIDMYGKCLKPHDARKVFDEMADSNEVT 142
AC G +H + +G+ + +LIDMY KC DAR VFD M T
Sbjct: 4 ACTEMGSLKLGRRVHDFALKNGFELEPFLGTALIDMYSKCGNLDDARTVFDMMQMRTLAT 183
Query: 143 WCSLLFAYANSSLFGMALEVFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLY 202
W NTMI G + L +F+EM ++
Sbjct: 184 W-------------------------------NTMITSLGVHGYRDEALSIFEEMEKANE 270
Query: 203 QPDQWTFSALMNACTESRDM 222
PD TF +++AC D+
Sbjct: 271 VPDAITFVGVLSACVYMNDL 330
Score = 40.0 bits (92), Expect = 0.003
Identities = 33/134 (24%), Positives = 52/134 (38%)
Frame = +1
Query: 215 ACTESRDMLYGCMVHGFVIKSGWSSAMEVKNSILSFYAKLECPSDAMEMFNSFGAFNQVS 274
ACTE + G VH F +K+G+ +E F
Sbjct: 4 ACTEMGSLKLGRRVHDFALKNGFE----------------------LEPFLG-------- 93
Query: 275 WNAIIDAHMKLGDTQKAFLAFQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSI 334
A+ID + K G+ A F + + +W +MI +G + ALS+F +M + +
Sbjct: 94 -TALIDMYSKCGNLDDARTVFDMMQMRTLATWNTMITSLGVHGYRDEALSIFEEMEKANE 270
Query: 335 QLDNLVAGAVLHAC 348
D + VL AC
Sbjct: 271 VPDAITFVGVLSAC 312
>AW317686
Length = 447
Score = 101 bits (252), Expect = 9e-22
Identities = 49/140 (35%), Positives = 78/140 (55%)
Frame = +1
Query: 402 KDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAF 461
+D+V+WN+++ F +G E++ +FR M+ + V+P+ VTF G+L C+H GL +EG
Sbjct: 28 RDVVTWNTLITGFAQNGHGEESLAVFRRMIEAKVEPNHVTFLGVLSGCNHAGLDNEGLQL 207
Query: 462 FRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTSGARTNSYEVLLGACHA 521
M ++G+ +H A ++D+LGR + EA SL +K + +LGAC
Sbjct: 208 VDLMERQYGVKPKAEHYALLIDLLGRRNRLMEAMSLIEKVPDGIKNHIAVWGAVLGACXV 387
Query: 522 HGDLGTGSSVGEYLKTLEPE 541
HG+L E L LEPE
Sbjct: 388 HGNLDLARKAAEKLFELEPE 447
Score = 45.8 bits (107), Expect = 6e-05
Identities = 19/58 (32%), Positives = 32/58 (54%)
Frame = +1
Query: 162 VFRSMPERVEIAWNTMIAGHARRGEVEACLGLFKEMCESLYQPDQWTFSALMNACTES 219
+F P R + WNT+I G A+ G E L +F+ M E+ +P+ TF +++ C +
Sbjct: 7 LFEMAPMRDVVTWNTLITGFAQNGHGEESLAVFRRMIEAKVEPNHVTFLGVLSGCNHA 180
Score = 43.9 bits (102), Expect = 2e-04
Identities = 17/54 (31%), Positives = 35/54 (64%)
Frame = +1
Query: 295 FQQAPDKNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHAC 348
F+ AP +++V+W ++I G+ +NG+GE +L++F M ++ +++ VL C
Sbjct: 10 FEMAPMRDVVTWNTLITGFAQNGHGEESLAVFRRMIEAKVEPNHVTFLGVLSGC 171
>TC222432
Length = 951
Score = 61.6 bits (148), Expect(2) = 1e-21
Identities = 40/129 (31%), Positives = 64/129 (49%), Gaps = 1/129 (0%)
Frame = +2
Query: 469 FGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYS-KTSGARTNSYEVLLGACHAHGDLGT 527
FG + ++H CMVD+LGR G + EA++L + +G +S+ L AC D+
Sbjct: 200 FGFAPQVEHYGCMVDLLGRAGCLDEAENLIQTMPYDANGIILSSF---LFACGYFNDVLR 370
Query: 528 GSSVGEYLKTLEPEKEVGYVMLSNLYCASGQWKEAEIVRKEMLDQGVKKVPGSSWIEIRN 587
V + + ++ + YVML NLY +W + E V++ M +G K S IEI
Sbjct: 371 AERVLKEVVKMDEDVAGNYVMLRNLYATRQRWTDVEDVKQMMKKRGTSKEVACSVIEIGG 550
Query: 588 VVTAFVSGN 596
F +G+
Sbjct: 551 SFIEFAAGD 577
Score = 60.5 bits (145), Expect(2) = 1e-21
Identities = 25/63 (39%), Positives = 41/63 (64%)
Frame = +3
Query: 403 DLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAFF 462
+ SWN+++ F ++G A EA+ +F M+ G P+EVT G+L C+H GL++EG +F
Sbjct: 3 ETASWNALINGFAVNGCAKEALEVFARMIEEGFGPNEVTMIGVLSACNHCGLVEEGRRWF 182
Query: 463 RSM 465
+M
Sbjct: 183 NAM 191
>BI893521
Length = 423
Score = 99.8 bits (247), Expect = 3e-21
Identities = 51/127 (40%), Positives = 73/127 (57%)
Frame = +1
Query: 402 KDLVSWNSMLFAFGLHGRANEAMCLFREMVASGVKPDEVTFTGMLMTCSHLGLIDEGFAF 461
+++++W S++ F HG A +A+ LF EM+ GVKP+EVT+ +L CSH+GLIDE +
Sbjct: 25 RNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSACSHVGLIDEAWKH 204
Query: 462 FRSMSSEFGLSHGMDHVACMVDMLGRGGYVAEAQSLAKKYSKTSGARTNSYEVLLGACHA 521
F SM +S M+H ACMVD+LGR G + EA S A + LG+C
Sbjct: 205 FNSMHYNHSISPRMEHYACMVDLLGRSGLLLEAIEFIN--SMPFDADALVWRTFLGSCRV 378
Query: 522 HGDLGTG 528
H + G
Sbjct: 379 HRNTKLG 399
Score = 40.8 bits (94), Expect = 0.002
Identities = 16/54 (29%), Positives = 34/54 (62%)
Frame = +1
Query: 301 KNIVSWTSMIVGYTRNGNGELALSMFLDMTRNSIQLDNLVAGAVLHACASLAIL 354
+N+++WTS+I G+ ++G AL +F +M ++ + + AVL AC+ + ++
Sbjct: 25 RNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSACSHVGLI 186
Score = 40.0 bits (92), Expect = 0.003
Identities = 18/57 (31%), Positives = 34/57 (59%)
Frame = +1
Query: 29 KLFDEMPDRDSVAWNAMITAYSHLGLYQQSLSLFGSMRISNSKPDSFSYSAALSACA 85
++F++M R+ + W ++I+ ++ G ++L LF M KP+ +Y A LSAC+
Sbjct: 1 QVFNDMGYRNVITWTSIISGFAKHGFATKALELFYEMLEIGVKPNEVTYIAVLSACS 171
Score = 31.2 bits (69), Expect = 1.5
Identities = 14/39 (35%), Positives = 22/39 (55%)
Frame = +1
Query: 130 KVFDEMADSNEVTWCSLLFAYANSSLFGMALEVFRSMPE 168
+VF++M N +TW S++ +A ALE+F M E
Sbjct: 1 QVFNDMGYRNVITWTSIISGFAKHGFATKALELFYEMLE 117
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.134 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,614,216
Number of Sequences: 63676
Number of extensions: 375661
Number of successful extensions: 1979
Number of sequences better than 10.0: 160
Number of HSP's better than 10.0 without gapping: 1705
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1924
length of query: 630
length of database: 12,639,632
effective HSP length: 103
effective length of query: 527
effective length of database: 6,081,004
effective search space: 3204689108
effective search space used: 3204689108
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0238.21


