

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
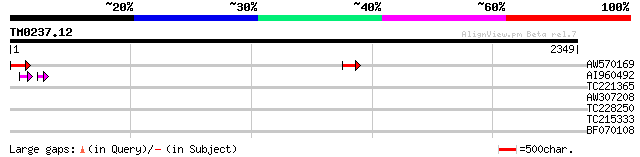
Query= TM0237.12
(2349 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW570169 149 2e-35
AI960492 38 8e-06
TC221365 similar to UP|Q96W55 (Q96W55) Ribosomal protein L39, co... 33 1.9
AW307208 similar to GP|20804893|dbj proline transport protein-li... 32 3.2
TC228250 UP|Q84P29 (Q84P29) Seed calcium dependent protein kinas... 32 4.2
TC215333 similar to UP|P93344 (P93344) Aldehyde dehydrogenase (N... 31 7.2
BF070108 31 7.2
>AW570169
Length = 366
Score = 149 bits (375), Expect = 2e-35
Identities = 74/80 (92%), Positives = 76/80 (94%)
Frame = +2
Query: 5 SPESIFLEDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHAG 64
SPESIFLEDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSH
Sbjct: 125 SPESIFLEDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHPA 304
Query: 65 DSLLSSSLSQWQGPALLAYN 84
SLLS SL+QWQGPALLA+N
Sbjct: 305 GSLLSDSLAQWQGPALLAFN 364
Score = 70.9 bits (172), Expect = 6e-12
Identities = 35/73 (47%), Positives = 46/73 (62%)
Frame = +2
Query: 1380 EAFGQHEALTTRLKHILEMYADGPGTLFELVQNAEDAGASEVIFLLDKSQYGTSSVLSPE 1439
E FGQ LT R++ +L Y +G L EL+QNA+DAGA+ V LD+ + S+LS
Sbjct: 146 EDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHPAGSLLSDS 325
Query: 1440 MADWQGPALYCFN 1452
+A WQGPAL FN
Sbjct: 326 LAQWQGPALLAFN 364
>AI960492
Length = 391
Score = 37.7 bits (86), Expect(2) = 8e-06
Identities = 20/55 (36%), Positives = 33/55 (59%), Gaps = 1/55 (1%)
Frame = +2
Query: 40 ELIQNADDAGATTVSLCLDRRSHAGDSLLSSSLSQWQGPALLA-YNDAVFTEDDF 93
+L++ AD A + L D+R H SLL +L ++QGPAL+A + A + ++F
Sbjct: 8 DLLELADCCKAKRLHLIYDKREHPRQSLLQHNLGEFQGPALVAIFECACLSREEF 172
Score = 32.3 bits (72), Expect(2) = 8e-06
Identities = 16/44 (36%), Positives = 23/44 (51%)
Frame = +3
Query: 115 GVGFNSVYHLTDLPSFVSGKYVVLFDPQGVYLPRVSAANPGKRI 158
G+G Y + DL S +SG Y +FDP+G L S P ++
Sbjct: 228 GLGLVCCYSICDLLSVISGGYFYMFDPRG*VLGAPSTNPPSAKM 359
>TC221365 similar to UP|Q96W55 (Q96W55) Ribosomal protein L39, complete
Length = 514
Score = 32.7 bits (73), Expect = 1.9
Identities = 12/25 (48%), Positives = 15/25 (60%)
Frame = -1
Query: 1113 MRILDGECSSTALLYCLGWMCPPGG 1137
+R DG C+S + C GW CP GG
Sbjct: 199 LRCRDGSCASASASRCSGWCCPFGG 125
>AW307208 similar to GP|20804893|dbj proline transport protein-like {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 397
Score = 32.0 bits (71), Expect = 3.2
Identities = 19/45 (42%), Positives = 25/45 (55%)
Frame = -3
Query: 966 PSGVFRESETLDIMRGLGLKTSVSPDTVLESARCIEHLMHEDQQK 1010
PSG + E L+I L L TS+SP ESA I+ HE ++K
Sbjct: 392 PSGHMTKLEKLNIHPQLKLVTSISPTCCHESADGIDQFHHEKKKK 258
>TC228250 UP|Q84P29 (Q84P29) Seed calcium dependent protein kinase a,
complete
Length = 1843
Score = 31.6 bits (70), Expect = 4.2
Identities = 27/101 (26%), Positives = 46/101 (44%), Gaps = 3/101 (2%)
Frame = +3
Query: 516 LFDMLL-KYNSSKVITPGTVRQFLRECESCNHLSRAHKLLLLEYCLEDLVDDDVGKAA-- 572
LFD ++ K + S+ ++ + E+C+ L H+ L E L D +D+D A
Sbjct: 480 LFDRIVQKGHYSERQAARLIKTIVEVVEACHSLGVMHRDLKPENFLFDTIDEDAKLKATD 659
Query: 573 YNLPLLPLANGNFASFLEASKGIPYFICDELEYKLLEPVSD 613
+ L + +F + G PY++ E+ KL P SD
Sbjct: 660 FGLSVFYKPGESFCDVV----GSPYYVAPEVLRKLYGPESD 770
>TC215333 similar to UP|P93344 (P93344) Aldehyde dehydrogenase (NAD+) ,
partial (73%)
Length = 1405
Score = 30.8 bits (68), Expect = 7.2
Identities = 22/60 (36%), Positives = 28/60 (46%), Gaps = 5/60 (8%)
Frame = +2
Query: 2038 HRKLCYIAQTDSTNISFLSC-----QLLEKLLVKLLPVEWQHASLVSWTPGIHGQPSLEW 2092
HR L Y+ Q + FL L +LLV LL W+ SL+ P I G+ SL W
Sbjct: 275 HRYLLYMQQNYFMRLDFLLVF*M*YLALVQLLVLLLLATWKLTSLLLLVPLILGKLSLNW 454
>BF070108
Length = 416
Score = 30.8 bits (68), Expect = 7.2
Identities = 22/92 (23%), Positives = 41/92 (43%)
Frame = -3
Query: 467 APVIYSNLGGGRWVSPSEAFLHDEKFTKSKDLSLALMQLGMPVVHLPNSLFDMLLKYNSS 526
A +I SN P +A LHD + D ++ + L +P++ +PN+ + ++
Sbjct: 261 ASIILSNF-------PVQAKLHDSSANPTLDCLVSYITLWVPLMIIPNNSSVICIRKTCK 103
Query: 527 KVITPGTVRQFLRECESCNHLSRAHKLLLLEY 558
+ ++ EC SC HL + L + Y
Sbjct: 102 RQADLRSILFHNVECASCIHLQLSIALKMHTY 7
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 109,524,388
Number of Sequences: 63676
Number of extensions: 1668624
Number of successful extensions: 7681
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 7413
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7678
length of query: 2349
length of database: 12,639,632
effective HSP length: 113
effective length of query: 2236
effective length of database: 5,444,244
effective search space: 12173329584
effective search space used: 12173329584
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0237.12


